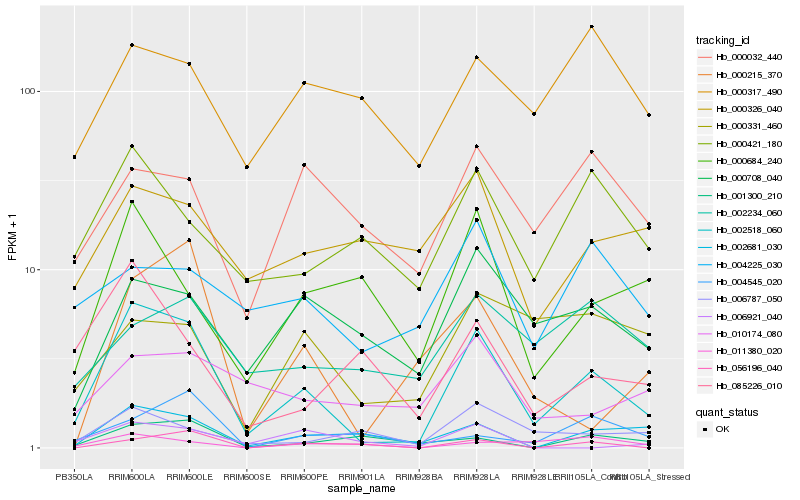
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002518_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631129 [Jatropha curcas] |
| 2 |
Hb_002681_030 |
0.2685478287 |
- |
- |
PREDICTED: fatty-acid-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_000215_370 |
0.2946180703 |
- |
- |
myoinositol oxygenase, putative [Ricinus communis] |
| 4 |
Hb_000421_180 |
0.2992366037 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000708_040 |
0.2993409752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_085226_010 |
0.302282684 |
- |
- |
hypothetical protein POPTR_0002s03930g [Populus trichocarpa] |
| 7 |
Hb_000684_240 |
0.3023318627 |
- |
- |
hypothetical protein RCOM_0819620 [Ricinus communis] |
| 8 |
Hb_006787_050 |
0.3051115404 |
- |
- |
hypothetical protein JCGZ_23988 [Jatropha curcas] |
| 9 |
Hb_001300_210 |
0.3091037619 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 23-like [Jatropha curcas] |
| 10 |
Hb_056196_040 |
0.3163615421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000032_440 |
0.3182243163 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_011380_020 |
0.3205333425 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X4 [Populus euphratica] |
| 13 |
Hb_010174_080 |
0.3205539927 |
- |
- |
- |
| 14 |
Hb_000326_040 |
0.3251427742 |
- |
- |
hypothetical protein PHAVU_006G187900g [Phaseolus vulgaris] |
| 15 |
Hb_004225_030 |
0.3255514555 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 16 |
Hb_002234_060 |
0.3264435714 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 17 |
Hb_004545_020 |
0.3278634595 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1-like [Populus euphratica] |
| 18 |
Hb_000317_490 |
0.3280247676 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 19 |
Hb_006921_040 |
0.3281743756 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 20 |
Hb_000331_460 |
0.3289277929 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |