| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
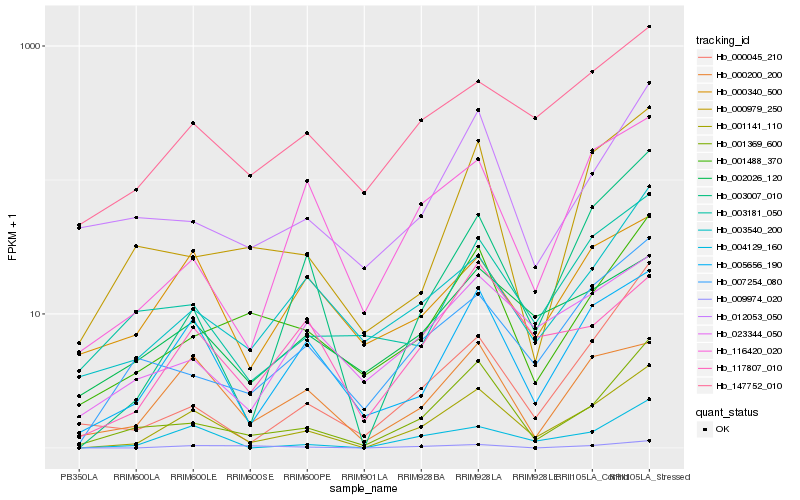
Hb_001141_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 2 |
Hb_004129_160 |
0.1486329324 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 3 |
Hb_005656_190 |
0.156447789 |
- |
- |
proteasome subunit beta type 6,9, putative [Ricinus communis] |
| 4 |
Hb_001369_600 |
0.1756979447 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000200_200 |
0.198615489 |
- |
- |
PREDICTED: putative zinc finger protein At1g68190 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003007_010 |
0.2024500991 |
- |
- |
PREDICTED: probable peptide/nitrate transporter At3g43790 isoform X1 [Populus euphratica] |
| 7 |
Hb_007254_080 |
0.2075017083 |
- |
- |
- |
| 8 |
Hb_147752_010 |
0.2081807571 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 9 |
Hb_001488_370 |
0.2133050125 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 10 |
Hb_003540_200 |
0.2177411184 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 11 |
Hb_117807_010 |
0.2179249356 |
- |
- |
PREDICTED: uncharacterized protein LOC105635720 [Jatropha curcas] |
| 12 |
Hb_009974_020 |
0.2245456983 |
- |
- |
PREDICTED: uncharacterized protein LOC104427753 [Eucalyptus grandis] |
| 13 |
Hb_116420_020 |
0.2249881962 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 14 |
Hb_003181_050 |
0.2250045283 |
- |
- |
PREDICTED: DNA replication complex GINS protein PSF2 [Jatropha curcas] |
| 15 |
Hb_002026_120 |
0.2294293764 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 16 |
Hb_000045_210 |
0.230217084 |
- |
- |
- |
| 17 |
Hb_000340_500 |
0.2306493419 |
- |
- |
PREDICTED: aldo-keto reductase-like [Jatropha curcas] |
| 18 |
Hb_023344_050 |
0.2307044533 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 19 |
Hb_012053_050 |
0.232548639 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 20 |
Hb_000979_250 |
0.2330545359 |
- |
- |
conserved hypothetical protein [Ricinus communis] |