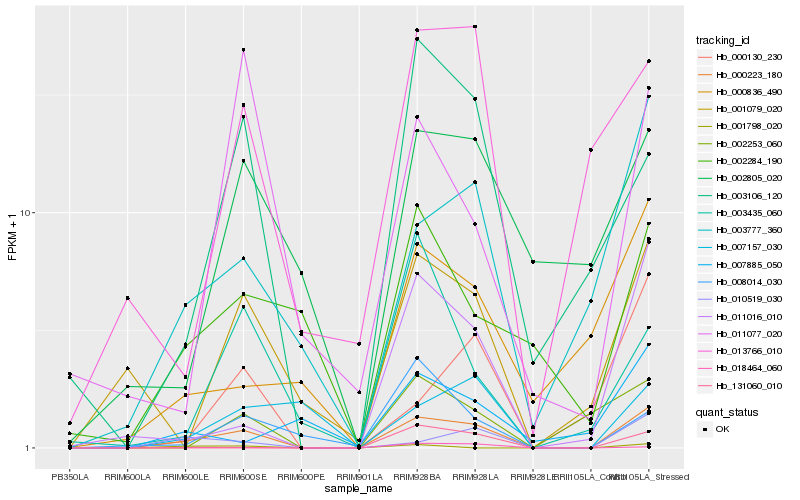
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000223_180 |
0.0 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 2 |
Hb_003777_360 |
0.2642641867 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 3 |
Hb_011016_010 |
0.265657068 |
- |
- |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 4 |
Hb_010519_030 |
0.2709522889 |
- |
- |
- |
| 5 |
Hb_001079_020 |
0.2863175309 |
- |
- |
hypothetical protein POPTR_0012s13720g [Populus trichocarpa] |
| 6 |
Hb_003106_120 |
0.3034558808 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 7 |
Hb_000130_230 |
0.3057458579 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 8 |
Hb_002253_060 |
0.3098848652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_013766_010 |
0.3129111025 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 10 |
Hb_000836_490 |
0.3170691288 |
- |
- |
PREDICTED: E3 SUMO-protein ligase pli1-like [Jatropha curcas] |
| 11 |
Hb_002284_190 |
0.3249136578 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 12 |
Hb_008014_030 |
0.3274782969 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 13 |
Hb_002805_020 |
0.3282323366 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_007157_030 |
0.3335155494 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 15 |
Hb_018464_060 |
0.3373555841 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_003435_060 |
0.3385624477 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 17 |
Hb_131060_010 |
0.3415455391 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 18 |
Hb_007885_050 |
0.3424855986 |
- |
- |
PREDICTED: lignin-forming anionic peroxidase-like [Jatropha curcas] |
| 19 |
Hb_001798_020 |
0.3434070531 |
- |
- |
PREDICTED: uncharacterized protein LOC104594032 [Nelumbo nucifera] |
| 20 |
Hb_011077_020 |
0.3451178896 |
- |
- |
kinase, putative [Ricinus communis] |