| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
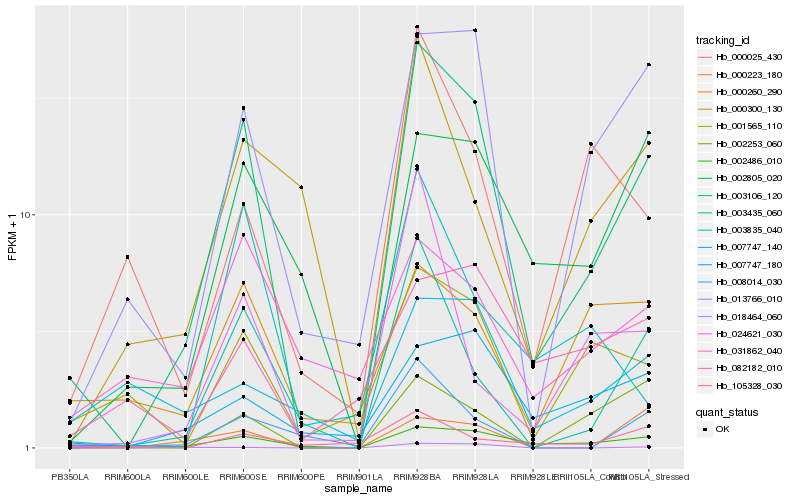
Hb_003106_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 2 |
Hb_002486_010 |
0.225145895 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 1 [Jatropha curcas] |
| 3 |
Hb_013766_010 |
0.2276758791 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 4 |
Hb_003435_060 |
0.2294123814 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 5 |
Hb_001565_110 |
0.2681207399 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 6 |
Hb_007747_180 |
0.2712848775 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 7 |
Hb_002253_060 |
0.2729255883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002805_020 |
0.2733106727 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_008014_030 |
0.2767469663 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 10 |
Hb_018464_060 |
0.284355015 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000260_290 |
0.2911082558 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000300_130 |
0.2938340446 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 13 |
Hb_024621_030 |
0.294069713 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 14 |
Hb_105328_030 |
0.2957714414 |
- |
- |
Putative retroelement [Oryza sativa Japonica Group] |
| 15 |
Hb_000025_430 |
0.2978566982 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000223_180 |
0.3034558808 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 17 |
Hb_003835_040 |
0.3040737684 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 18 |
Hb_082182_010 |
0.3059035929 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 19 |
Hb_031862_040 |
0.305975393 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 20 |
Hb_007747_140 |
0.3064000272 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |