| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
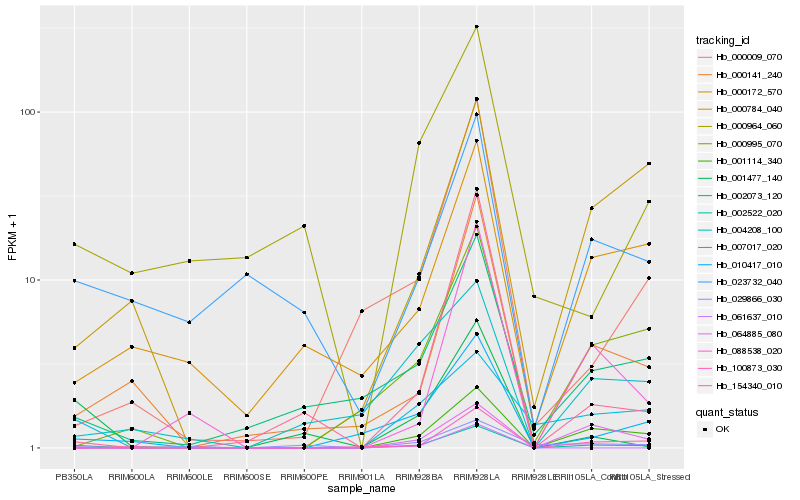
Hb_002522_020 |
0.0 |
- |
- |
hypothetical protein B456_011G148700, partial [Gossypium raimondii] |
| 2 |
Hb_088538_020 |
0.244916394 |
- |
- |
hypothetical protein JCGZ_12178 [Jatropha curcas] |
| 3 |
Hb_001114_340 |
0.247892847 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 4 |
Hb_010417_010 |
0.2497923685 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000141_240 |
0.2564701488 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002073_120 |
0.2568088499 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein [Theobroma cacao] |
| 7 |
Hb_000172_570 |
0.2663991619 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 8 |
Hb_023732_040 |
0.2715150654 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_000995_070 |
0.2724487359 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 10 |
Hb_001477_140 |
0.2759122337 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_007017_020 |
0.2761963683 |
- |
- |
hypothetical protein JCGZ_21740 [Jatropha curcas] |
| 12 |
Hb_100873_030 |
0.2788320303 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 13 |
Hb_000964_060 |
0.2881718239 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 14 |
Hb_000009_070 |
0.2906757404 |
- |
- |
PREDICTED: globulin-1 S allele [Jatropha curcas] |
| 15 |
Hb_004208_100 |
0.2960608825 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 16 |
Hb_064885_080 |
0.2963948493 |
- |
- |
- |
| 17 |
Hb_154340_010 |
0.3000123756 |
- |
- |
truncated nitrate transporter [Fragaria vesca] |
| 18 |
Hb_000784_040 |
0.3053367256 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 19 |
Hb_029866_030 |
0.3077768093 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 20 |
Hb_061637_010 |
0.3085389492 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |