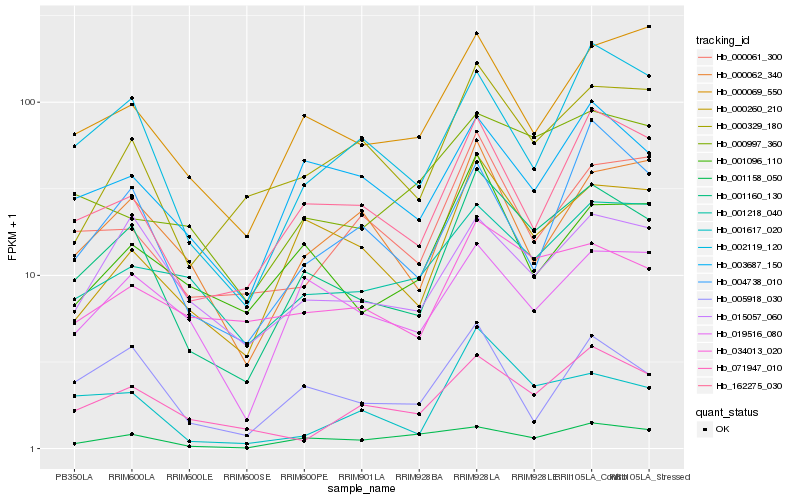
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001160_130 |
0.0 |
- |
- |
Glycolipid transfer protein, putative [Ricinus communis] |
| 2 |
Hb_162275_030 |
0.1417313829 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 3 |
Hb_000260_210 |
0.1462833793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003687_150 |
0.1518515346 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase XBAT35 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002119_120 |
0.1538704326 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_015057_060 |
0.1548190769 |
- |
- |
- |
| 7 |
Hb_000329_180 |
0.1597921507 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 8 |
Hb_034013_020 |
0.1599641687 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 9 |
Hb_000069_550 |
0.1605573818 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 10 |
Hb_001617_020 |
0.163038073 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_019516_080 |
0.168276239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001096_110 |
0.1689105248 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 13 |
Hb_005918_030 |
0.1694550546 |
- |
- |
hypothetical protein JCGZ_24769 [Jatropha curcas] |
| 14 |
Hb_001158_050 |
0.1696909069 |
- |
- |
- |
| 15 |
Hb_071947_010 |
0.1697582604 |
- |
- |
- |
| 16 |
Hb_000062_340 |
0.171320631 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 17 |
Hb_000997_360 |
0.1725091036 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |
| 18 |
Hb_001218_040 |
0.1741813555 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 19 |
Hb_000061_300 |
0.1743669544 |
- |
- |
hypothetical protein JCGZ_11668 [Jatropha curcas] |
| 20 |
Hb_004738_010 |
0.1756547749 |
- |
- |
PREDICTED: N-acetyl-D-glucosamine kinase-like [Jatropha curcas] |