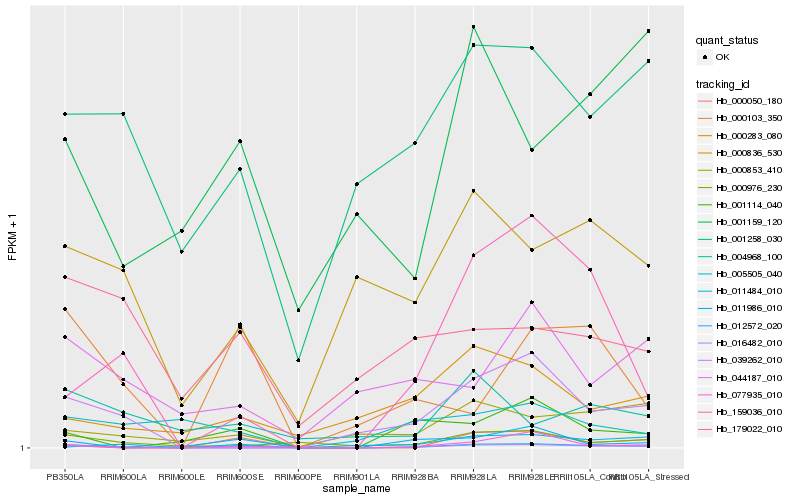
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012572_020 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_016482_010 |
0.2482591367 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000103_350 |
0.2739589653 |
- |
- |
- |
| 4 |
Hb_159036_010 |
0.2788239987 |
- |
- |
- |
| 5 |
Hb_039262_010 |
0.2887882535 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 6 |
Hb_011986_010 |
0.2893934823 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_000853_410 |
0.2924120499 |
- |
- |
- |
| 8 |
Hb_001258_030 |
0.2928571537 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 9 |
Hb_000836_530 |
0.2942175087 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 10 |
Hb_000976_230 |
0.2948275271 |
- |
- |
PREDICTED: probable terpene synthase 4 [Jatropha curcas] |
| 11 |
Hb_011484_010 |
0.3012966251 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_001114_040 |
0.3047786268 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 13 |
Hb_004968_100 |
0.3055134116 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 14 |
Hb_000050_180 |
0.3105872808 |
- |
- |
- |
| 15 |
Hb_179022_010 |
0.3151386581 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_000283_080 |
0.3152889961 |
- |
- |
- |
| 17 |
Hb_044187_010 |
0.3200903543 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 18 |
Hb_001159_120 |
0.322058075 |
- |
- |
PREDICTED: uncharacterized protein LOC105645945 [Jatropha curcas] |
| 19 |
Hb_005505_040 |
0.3223318725 |
- |
- |
- |
| 20 |
Hb_077935_010 |
0.3233992194 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |