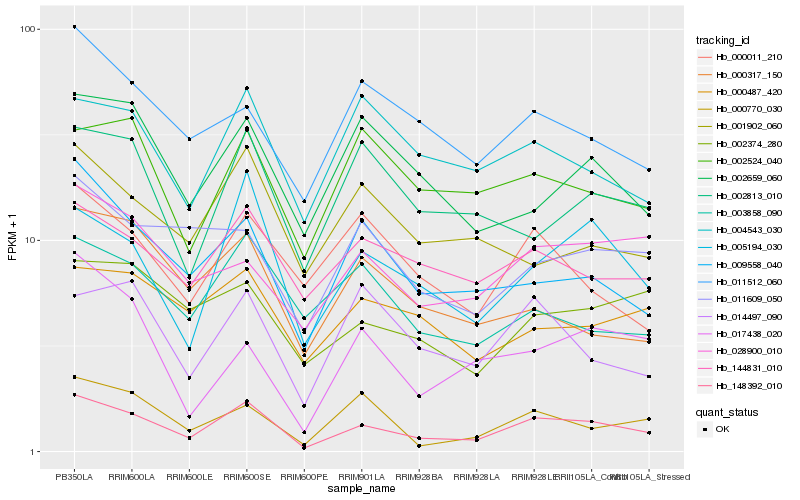
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148392_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_005194_030 |
0.1309955498 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009558_040 |
0.1546669906 |
transcription factor |
TF Family: MYB |
hypothetical protein PHAVU_008G102000g [Phaseolus vulgaris] |
| 4 |
Hb_144831_010 |
0.165005037 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000770_030 |
0.1677980492 |
- |
- |
- |
| 6 |
Hb_028900_010 |
0.1691384994 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 7 |
Hb_014497_090 |
0.1704517824 |
- |
- |
hypothetical protein PHAVU_002G037900g [Phaseolus vulgaris] |
| 8 |
Hb_002813_010 |
0.1709064516 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002374_280 |
0.1719746335 |
- |
- |
- |
| 10 |
Hb_004543_030 |
0.1723931791 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 11 |
Hb_002524_040 |
0.1726228802 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 12 |
Hb_002659_060 |
0.1730499638 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 13 |
Hb_011512_060 |
0.1733991753 |
- |
- |
PREDICTED: chaperonin CPN60-2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_003858_090 |
0.1759023578 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 15 |
Hb_000011_210 |
0.1762093268 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 16 |
Hb_011609_050 |
0.176731968 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_017438_020 |
0.1768100507 |
- |
- |
- |
| 18 |
Hb_001902_060 |
0.1783157771 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000487_420 |
0.178509167 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 20 |
Hb_000317_150 |
0.1792338672 |
- |
- |
PREDICTED: uncharacterized protein LOC104901272 [Beta vulgaris subsp. vulgaris] |