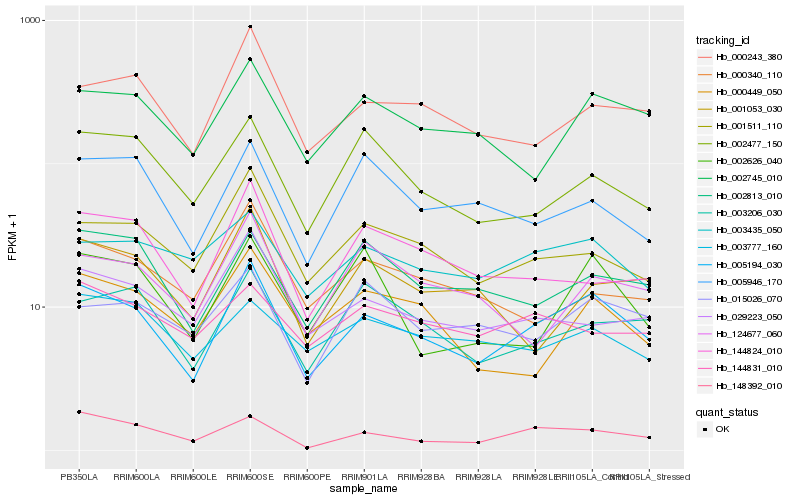
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005194_030 |
0.0 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 2 |
Hb_148392_010 |
0.1309955498 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_002745_010 |
0.1385668211 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 4 |
Hb_000243_380 |
0.1386285456 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 5 |
Hb_001511_110 |
0.1407487995 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_029223_050 |
0.1485767932 |
- |
- |
PREDICTED: uncharacterized protein LOC105642267 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003206_030 |
0.1502503937 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 8 |
Hb_002813_010 |
0.1504512818 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003435_050 |
0.1504845087 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1-like [Jatropha curcas] |
| 10 |
Hb_002477_150 |
0.1562356018 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_144824_010 |
0.1565767047 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 12 |
Hb_000449_050 |
0.1566190708 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 13 |
Hb_015026_070 |
0.1583727597 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005946_170 |
0.160272899 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 15 |
Hb_002626_040 |
0.1617397611 |
- |
- |
PREDICTED: inter-alpha-trypsin inhibitor heavy chain H3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_124677_060 |
0.1627738703 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_003777_160 |
0.1637379085 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000340_110 |
0.1643278018 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 19 |
Hb_001053_030 |
0.1644340396 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_144831_010 |
0.1655474651 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |