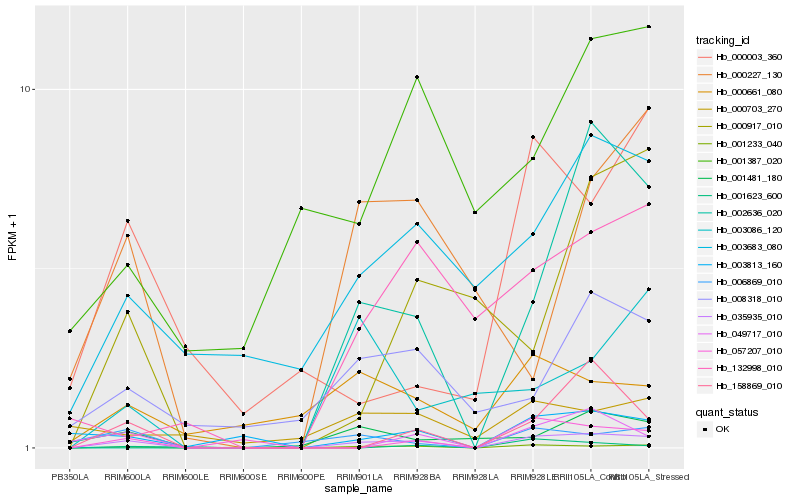
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035935_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 2 |
Hb_001233_040 |
0.2178331995 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 3 |
Hb_006869_010 |
0.3027997541 |
- |
- |
unnamed protein product [Coffea canephora] |
| 4 |
Hb_003813_160 |
0.3092996841 |
- |
- |
- |
| 5 |
Hb_158869_010 |
0.3099455669 |
- |
- |
PREDICTED: uncharacterized protein LOC105778995 [Gossypium raimondii] |
| 6 |
Hb_049717_010 |
0.3140900391 |
- |
- |
PREDICTED: uncharacterized protein LOC105800955 [Gossypium raimondii] |
| 7 |
Hb_003683_080 |
0.3181324109 |
- |
- |
Angio-associated migratory cell protein, putative [Ricinus communis] |
| 8 |
Hb_001481_180 |
0.3181811557 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 9 |
Hb_002636_020 |
0.32032996 |
- |
- |
- |
| 10 |
Hb_008318_010 |
0.3276657246 |
- |
- |
PREDICTED: WD repeat-containing protein 82-B isoform X1 [Jatropha curcas] |
| 11 |
Hb_057207_010 |
0.3281056242 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 12 |
Hb_000661_080 |
0.334997641 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 13 |
Hb_001623_600 |
0.3395289601 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 14 |
Hb_132998_010 |
0.3490800334 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 15 |
Hb_003086_120 |
0.3534304056 |
- |
- |
PREDICTED: uncharacterized protein LOC105628509 [Jatropha curcas] |
| 16 |
Hb_000227_130 |
0.3543774104 |
- |
- |
PREDICTED: peroxiredoxin-2F, mitochondrial [Jatropha curcas] |
| 17 |
Hb_001387_020 |
0.3593619056 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 18 |
Hb_000917_010 |
0.3599186702 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000003_360 |
0.3600616393 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 20 |
Hb_000703_270 |
0.3619435077 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |