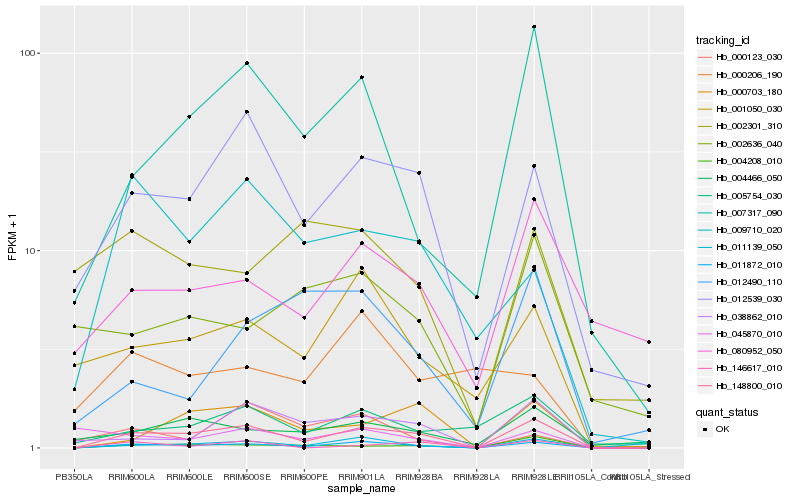
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011872_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_148800_010 |
0.1046732824 |
- |
- |
- |
| 3 |
Hb_012539_030 |
0.238756464 |
- |
- |
PREDICTED: uncharacterized protein LOC105638247 [Jatropha curcas] |
| 4 |
Hb_001050_030 |
0.2432779335 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 5 |
Hb_007317_090 |
0.2512894504 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 6 |
Hb_011139_050 |
0.2703135177 |
- |
- |
nicotinate phosphoribosyltransferase family protein [Populus trichocarpa] |
| 7 |
Hb_038862_010 |
0.2786339957 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_004208_010 |
0.283159156 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 9 |
Hb_009710_020 |
0.2907044501 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 10 |
Hb_005754_030 |
0.2912393509 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 11 |
Hb_004466_050 |
0.291715333 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 12 |
Hb_000703_180 |
0.2975817905 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |
| 13 |
Hb_146617_010 |
0.2977547912 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 14 |
Hb_012490_110 |
0.3020736117 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 15 |
Hb_000123_030 |
0.3032434303 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_080952_050 |
0.3046005939 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 17 |
Hb_000206_190 |
0.3109656499 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 18 |
Hb_002636_040 |
0.3185728888 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002301_310 |
0.3202132525 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.4 isoform X1 [Jatropha curcas] |
| 20 |
Hb_045870_010 |
0.3227415722 |
- |
- |
- |