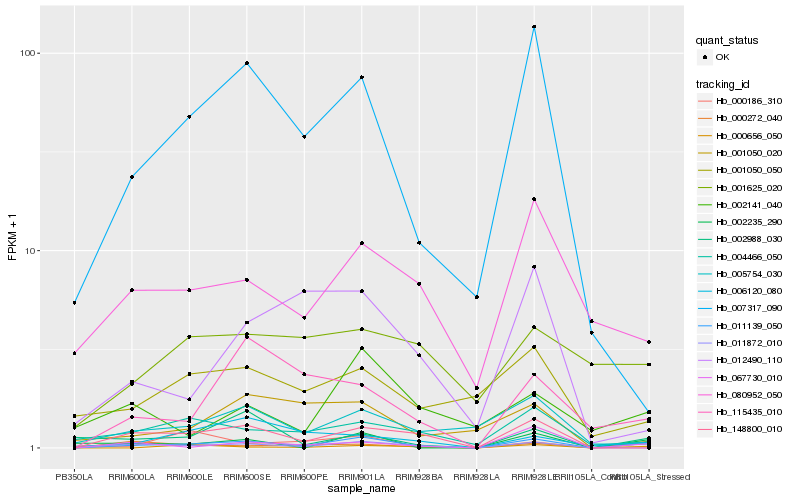
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011139_050 |
0.0 |
- |
- |
nicotinate phosphoribosyltransferase family protein [Populus trichocarpa] |
| 2 |
Hb_011872_010 |
0.2703135177 |
- |
- |
- |
| 3 |
Hb_000272_040 |
0.2869402141 |
- |
- |
- |
| 4 |
Hb_148800_010 |
0.2884044081 |
- |
- |
- |
| 5 |
Hb_000656_050 |
0.2914035741 |
- |
- |
PREDICTED: 3-oxoacyl-[acyl-carrier-protein] synthase II, chloroplastic-like [Vitis vinifera] |
| 6 |
Hb_007317_090 |
0.2939168856 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 7 |
Hb_004466_050 |
0.2994881028 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 8 |
Hb_080952_050 |
0.3002569575 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Nelumbo nucifera] |
| 9 |
Hb_002235_290 |
0.3003251324 |
- |
- |
PREDICTED: casparian strip membrane protein 5 [Jatropha curcas] |
| 10 |
Hb_115435_010 |
0.3020629388 |
- |
- |
- |
| 11 |
Hb_067730_010 |
0.3025992847 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |
| 12 |
Hb_005754_030 |
0.3115238714 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 13 |
Hb_001050_050 |
0.3188233734 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g57250, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002988_030 |
0.319760143 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 15 |
Hb_002141_040 |
0.3198266688 |
- |
- |
- |
| 16 |
Hb_006120_080 |
0.321987365 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |
| 17 |
Hb_001050_020 |
0.3240610983 |
- |
- |
- |
| 18 |
Hb_012490_110 |
0.3257572599 |
- |
- |
PREDICTED: uncharacterized membrane protein C776.05 [Jatropha curcas] |
| 19 |
Hb_001625_020 |
0.3269834041 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 20 |
Hb_000186_310 |
0.3284878138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |