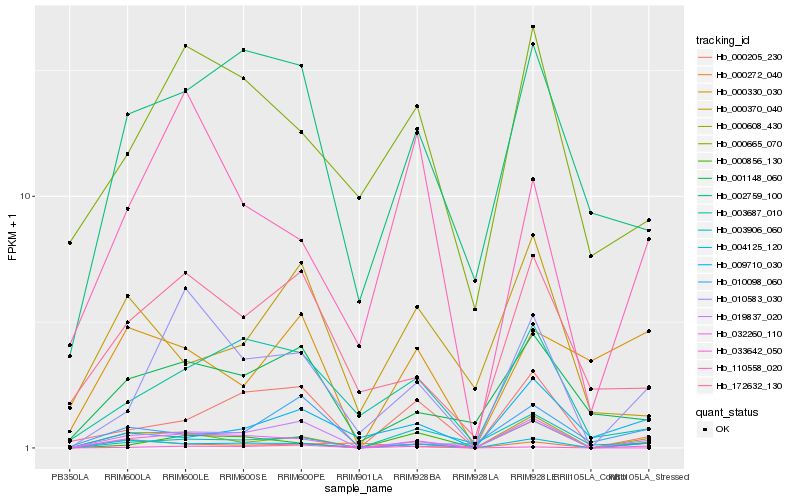
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004125_120 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_000608_430 |
0.1870405924 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_019837_020 |
0.2051863827 |
- |
- |
- |
| 4 |
Hb_000330_030 |
0.2807879269 |
- |
- |
PREDICTED: uncharacterized protein LOC105633814 [Jatropha curcas] |
| 5 |
Hb_002759_100 |
0.2854614548 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 6 |
Hb_000205_230 |
0.2927224174 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000856_130 |
0.2932263988 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_032260_110 |
0.2967881606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000272_040 |
0.2999391002 |
- |
- |
- |
| 10 |
Hb_001148_060 |
0.3025171844 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 11 |
Hb_033642_050 |
0.3051942218 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter B family member 4-like [Jatropha curcas] |
| 12 |
Hb_009710_030 |
0.3080360343 |
- |
- |
- |
| 13 |
Hb_010583_030 |
0.3099686642 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 14 |
Hb_003687_010 |
0.3131161852 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 15 |
Hb_172632_130 |
0.3177640218 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 16 |
Hb_000370_040 |
0.3237699842 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_010098_060 |
0.3242304804 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |
| 18 |
Hb_110558_020 |
0.327724447 |
- |
- |
PREDICTED: uncharacterized protein LOC105641876 [Jatropha curcas] |
| 19 |
Hb_003906_060 |
0.3286790716 |
- |
- |
hypothetical protein VITISV_022975 [Vitis vinifera] |
| 20 |
Hb_000665_070 |
0.3298576008 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3 isoform X1 [Jatropha curcas] |