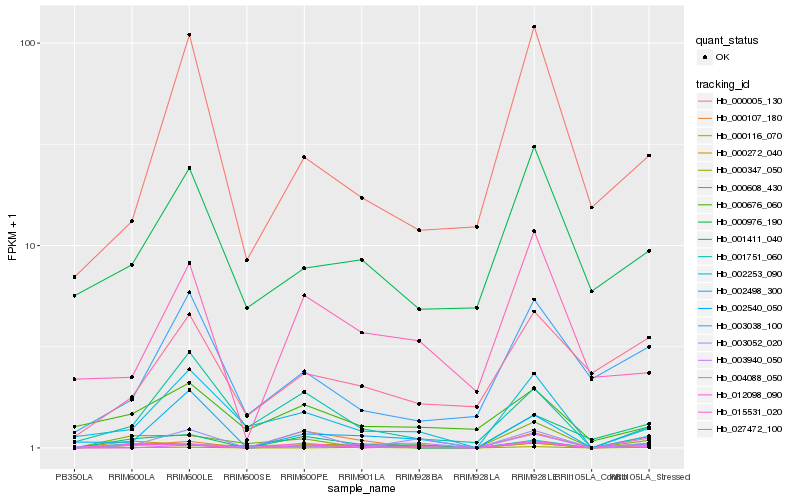
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000116_070 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 2 |
Hb_002253_090 |
0.2056779107 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_002540_050 |
0.2711586361 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000608_430 |
0.2741608641 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_000272_040 |
0.2775678582 |
- |
- |
- |
| 6 |
Hb_001411_040 |
0.2802276649 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 7 |
Hb_004088_050 |
0.2985633878 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 8 |
Hb_002498_300 |
0.3067724476 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 9 |
Hb_012098_090 |
0.3093971695 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_015531_020 |
0.3109776893 |
- |
- |
PREDICTED: uncharacterized protein LOC100267182 isoform X2 [Vitis vinifera] |
| 11 |
Hb_001751_060 |
0.3136120155 |
- |
- |
PREDICTED: SEC12-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000107_180 |
0.3137485522 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 13 |
Hb_003940_050 |
0.3156334461 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 14 |
Hb_027472_100 |
0.3162204137 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 15 |
Hb_000005_130 |
0.3177190541 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 16 |
Hb_000347_050 |
0.3187861671 |
- |
- |
PREDICTED: ABC transporter B family member 3-like [Jatropha curcas] |
| 17 |
Hb_000676_060 |
0.3214835679 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 18 |
Hb_000976_190 |
0.3220370146 |
- |
- |
PREDICTED: obg-like ATPase 1 [Jatropha curcas] |
| 19 |
Hb_003038_100 |
0.3224419052 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_003052_020 |
0.3225878742 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |