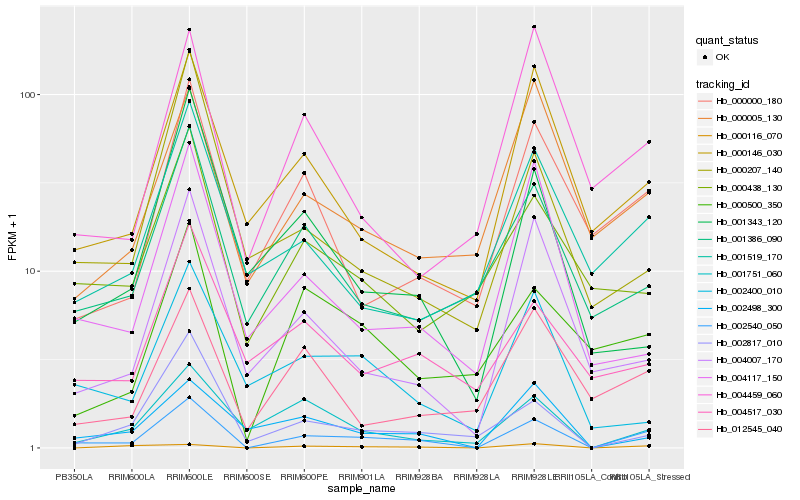
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002540_050 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Jatropha curcas] |
| 2 |
Hb_004007_170 |
0.2507319578 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 3 |
Hb_002817_010 |
0.2518785755 |
- |
- |
PREDICTED: uncharacterized protein LOC105630103 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001751_060 |
0.2524511471 |
- |
- |
PREDICTED: SEC12-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000146_030 |
0.2536981174 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002498_300 |
0.2538710681 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 7 |
Hb_000000_180 |
0.2580441449 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_000500_350 |
0.2586579891 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 9 |
Hb_001386_090 |
0.2591690608 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 10 |
Hb_001519_170 |
0.2621978018 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002400_010 |
0.2649354863 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 12 |
Hb_004117_150 |
0.2654037708 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 13 |
Hb_012545_040 |
0.2669912691 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000005_130 |
0.2680804556 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 15 |
Hb_000116_070 |
0.2711586361 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 16 |
Hb_001343_120 |
0.2718395459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000438_130 |
0.2720590427 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 18 |
Hb_000207_140 |
0.2733733602 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 19 |
Hb_004459_060 |
0.2740412801 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_004517_030 |
0.2744669604 |
- |
- |
conserved hypothetical protein [Ricinus communis] |