| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
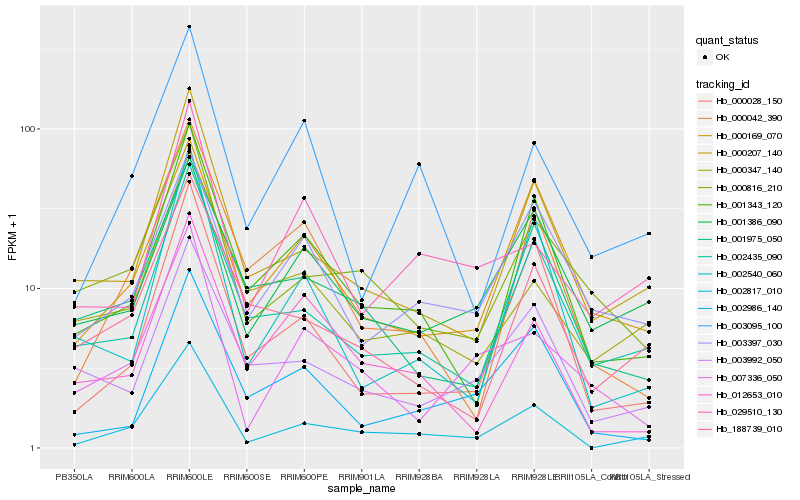
Hb_002817_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630103 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000207_140 |
0.1606428003 |
- |
- |
hypothetical protein POPTR_0019s11310g [Populus trichocarpa] |
| 3 |
Hb_001343_120 |
0.1720828161 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002435_090 |
0.1770978048 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000028_150 |
0.1808279921 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 6 |
Hb_000816_210 |
0.1924176851 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 7 |
Hb_002540_060 |
0.1929963245 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003095_100 |
0.1963737367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_188739_010 |
0.1969752329 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_000042_390 |
0.199813431 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 7 [Jatropha curcas] |
| 11 |
Hb_000347_140 |
0.2021235996 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_003992_050 |
0.202800954 |
- |
- |
delta 9 desaturase, putative [Ricinus communis] |
| 13 |
Hb_012653_010 |
0.2028453561 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 14 |
Hb_001386_090 |
0.2048163232 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 15 |
Hb_000169_070 |
0.2060812711 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 16 |
Hb_002986_140 |
0.2067294921 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 17 |
Hb_007336_050 |
0.2069595074 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 18 |
Hb_029510_130 |
0.2072766939 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 19 |
Hb_003397_030 |
0.2102551553 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001975_050 |
0.2125525082 |
- |
- |
Tyrosine-specific transport protein, putative [Ricinus communis] |