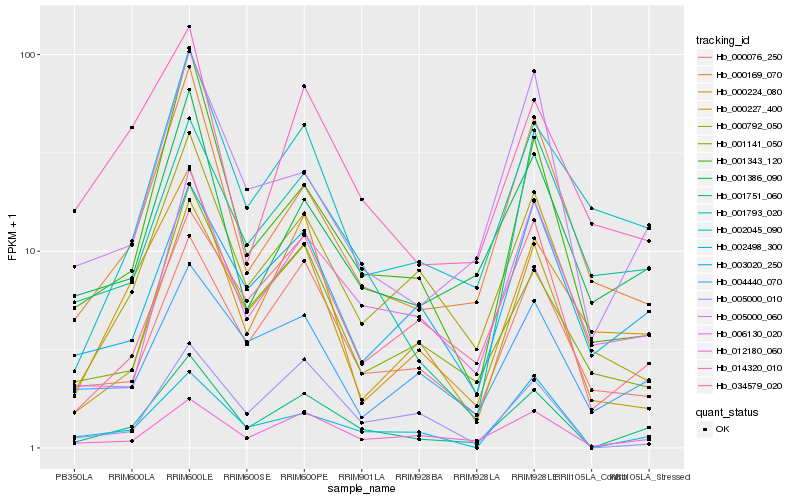
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001751_060 |
0.0 |
- |
- |
PREDICTED: SEC12-like protein 1 [Jatropha curcas] |
| 2 |
Hb_012180_060 |
0.1519178448 |
- |
- |
peptidase, putative [Ricinus communis] |
| 3 |
Hb_002498_300 |
0.1712895406 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 4 |
Hb_000076_250 |
0.1767027696 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005000_060 |
0.1813048711 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 6 |
Hb_000169_070 |
0.1828026971 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 7 |
Hb_001386_090 |
0.182926827 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 8 |
Hb_034579_020 |
0.1839542669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000792_050 |
0.1841266713 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_006130_020 |
0.1849689257 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003020_250 |
0.1871891348 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 12 |
Hb_001343_120 |
0.1874209995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001141_050 |
0.1879681755 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000227_400 |
0.1883896542 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 15 |
Hb_005000_010 |
0.190881887 |
- |
- |
PREDICTED: K(+) efflux antiporter 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001793_020 |
0.1911984907 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 17 |
Hb_002045_090 |
0.1915052925 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 18 |
Hb_000224_080 |
0.194198913 |
- |
- |
hypothetical protein RCOM_0510880 [Ricinus communis] |
| 19 |
Hb_004440_070 |
0.1944359099 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520 [Jatropha curcas] |
| 20 |
Hb_014320_010 |
0.1960722792 |
- |
- |
PREDICTED: uncharacterized protein LOC105629932 [Jatropha curcas] |