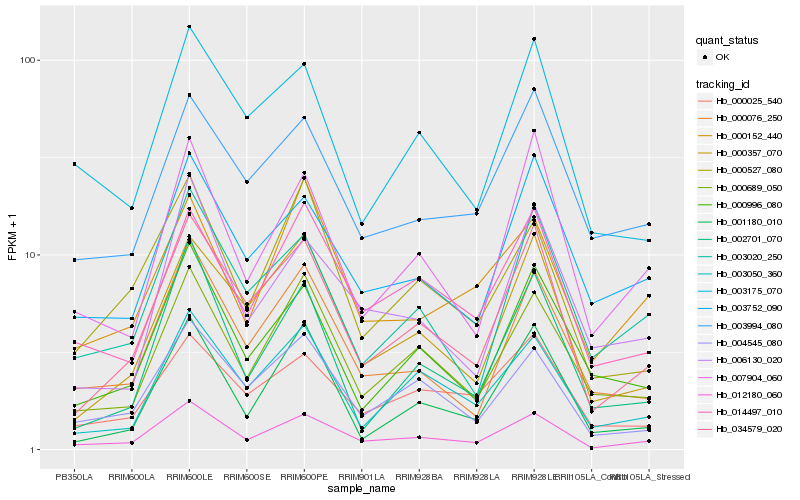
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012180_060 |
0.0 |
- |
- |
peptidase, putative [Ricinus communis] |
| 2 |
Hb_034579_020 |
0.0907093785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000689_050 |
0.1087209121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000076_250 |
0.1104186201 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004545_080 |
0.1126096056 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_007904_060 |
0.1137762787 |
- |
- |
PREDICTED: very-long-chain 3-oxoacyl-CoA reductase 1 [Jatropha curcas] |
| 7 |
Hb_014497_010 |
0.1203299544 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 8 |
Hb_003050_360 |
0.1207160055 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_003020_250 |
0.1217046319 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 10 |
Hb_000527_080 |
0.1243567875 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 11 |
Hb_000996_080 |
0.1246498573 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 12 |
Hb_003752_090 |
0.1255968534 |
- |
- |
chitinase, putative [Ricinus communis] |
| 13 |
Hb_000357_070 |
0.1262342973 |
- |
- |
- |
| 14 |
Hb_006130_020 |
0.1272824341 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_002701_070 |
0.1352989218 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 16 |
Hb_000025_540 |
0.1357591318 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 17 |
Hb_003175_070 |
0.1364818526 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 18 |
Hb_003994_080 |
0.1392847232 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 19 |
Hb_001180_010 |
0.1406504007 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 20 |
Hb_000152_440 |
0.140747487 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |