| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
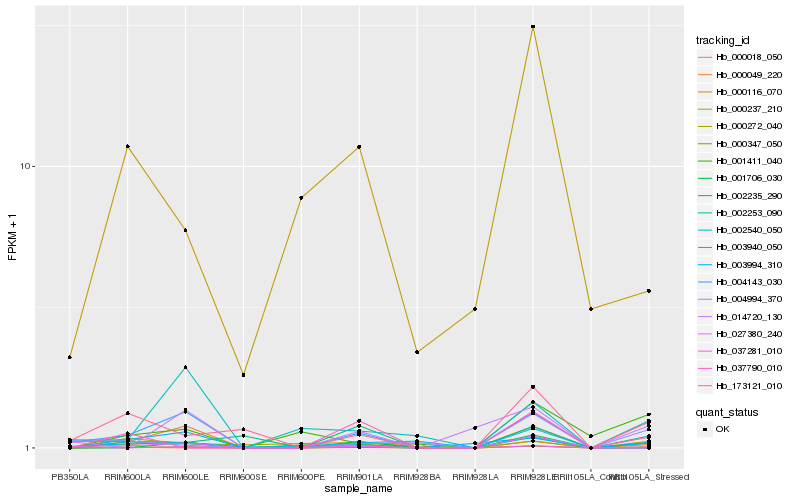
Hb_000347_050 |
0.0 |
- |
- |
PREDICTED: ABC transporter B family member 3-like [Jatropha curcas] |
| 2 |
Hb_002253_090 |
0.2366942207 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000116_070 |
0.3187861671 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 4 |
Hb_173121_010 |
0.3460120925 |
- |
- |
- |
| 5 |
Hb_000018_050 |
0.34658861 |
- |
- |
PREDICTED: uncharacterized protein LOC104899363 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_004143_030 |
0.3492471007 |
- |
- |
PREDICTED: chromosome-associated kinesin KIF4B [Jatropha curcas] |
| 7 |
Hb_000049_220 |
0.3519622343 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_037790_010 |
0.3550386906 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 9 |
Hb_003940_050 |
0.3582916018 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 10 |
Hb_003994_310 |
0.3675331008 |
transcription factor |
TF Family: PHD |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_027380_240 |
0.3841318506 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_037281_010 |
0.3844926183 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 13 |
Hb_001411_040 |
0.3905637125 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 14 |
Hb_014720_130 |
0.3917842575 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 15 |
Hb_002235_290 |
0.3938909643 |
- |
- |
PREDICTED: casparian strip membrane protein 5 [Jatropha curcas] |
| 16 |
Hb_004994_370 |
0.3942945832 |
- |
- |
- |
| 17 |
Hb_002540_050 |
0.3969352728 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g13770, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000237_210 |
0.3974833938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000272_040 |
0.3978001334 |
- |
- |
- |
| 20 |
Hb_001706_030 |
0.3980419821 |
- |
- |
PREDICTED: uncharacterized protein LOC104880364 [Vitis vinifera] |