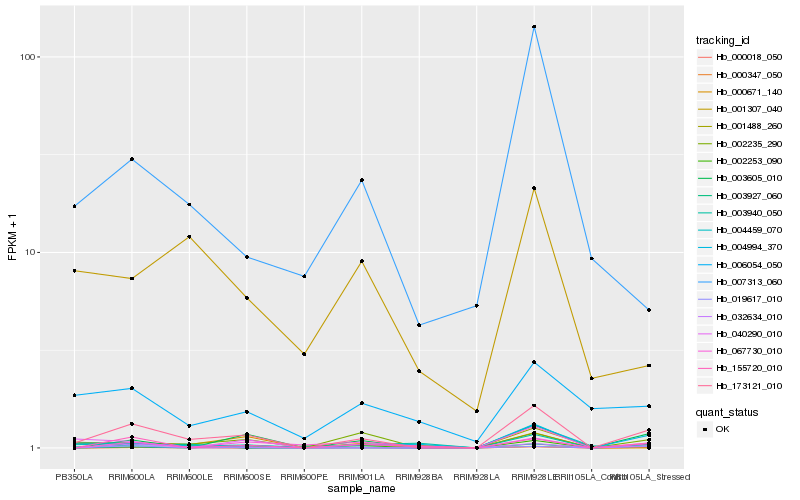
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_173121_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_032634_010 |
0.2544795334 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 3 |
Hb_003605_010 |
0.2816969538 |
- |
- |
- |
| 4 |
Hb_002235_290 |
0.288175528 |
- |
- |
PREDICTED: casparian strip membrane protein 5 [Jatropha curcas] |
| 5 |
Hb_019617_010 |
0.2980647844 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_067730_010 |
0.3157269495 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |
| 7 |
Hb_155720_010 |
0.3174243888 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 8 |
Hb_004459_070 |
0.3209988545 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 9 |
Hb_000671_140 |
0.3409727695 |
- |
- |
hypothetical protein VITISV_003767 [Vitis vinifera] |
| 10 |
Hb_000018_050 |
0.342731815 |
- |
- |
PREDICTED: uncharacterized protein LOC104899363 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_004994_370 |
0.3441084896 |
- |
- |
- |
| 12 |
Hb_007313_060 |
0.3448234751 |
- |
- |
PREDICTED: paramyosin [Jatropha curcas] |
| 13 |
Hb_000347_050 |
0.3460120925 |
- |
- |
PREDICTED: ABC transporter B family member 3-like [Jatropha curcas] |
| 14 |
Hb_040290_010 |
0.3466084784 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 15 |
Hb_002253_090 |
0.3509637034 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_006054_050 |
0.3509854883 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 17 |
Hb_001488_260 |
0.3524133932 |
- |
- |
Alpha-2-macroglobulin [Gossypium arboreum] |
| 18 |
Hb_003940_050 |
0.3527474791 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 19 |
Hb_001307_040 |
0.3574643672 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 4B [Jatropha curcas] |
| 20 |
Hb_003927_060 |
0.3578574369 |
- |
- |
hypothetical protein JCGZ_05636 [Jatropha curcas] |