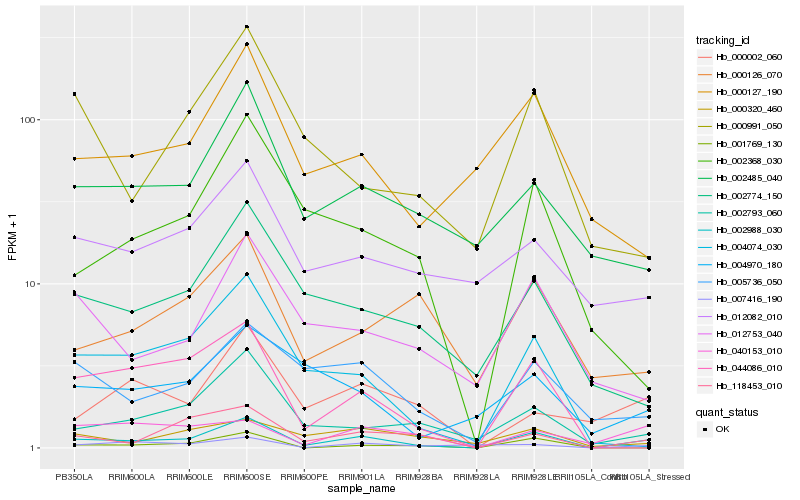
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002988_030 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 2 |
Hb_118453_010 |
0.2289974168 |
- |
- |
hypothetical protein VITISV_012360 [Vitis vinifera] |
| 3 |
Hb_002793_060 |
0.2369340151 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 4 |
Hb_005736_050 |
0.2413252109 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 5 |
Hb_044086_010 |
0.2430867323 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_004074_030 |
0.2467895328 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002485_040 |
0.2528661163 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 8 |
Hb_000002_060 |
0.2583290705 |
- |
- |
BnaA03g50800D [Brassica napus] |
| 9 |
Hb_000127_190 |
0.2635480893 |
- |
- |
PREDICTED: protein GIGANTEA [Jatropha curcas] |
| 10 |
Hb_002774_150 |
0.2637174271 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 11 |
Hb_012753_040 |
0.2648837781 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007416_190 |
0.2651526231 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 13 |
Hb_002368_030 |
0.2651793126 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 14 |
Hb_040153_010 |
0.2659293802 |
- |
- |
PREDICTED: DJ-1 protein homolog E-like [Jatropha curcas] |
| 15 |
Hb_001769_130 |
0.2661264137 |
- |
- |
- |
| 16 |
Hb_012082_010 |
0.2672646232 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004970_180 |
0.2689026572 |
- |
- |
- |
| 18 |
Hb_000320_460 |
0.2690183001 |
- |
- |
PREDICTED: uncharacterized protein LOC105650098 [Jatropha curcas] |
| 19 |
Hb_000991_050 |
0.2703040751 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 20 |
Hb_000126_070 |
0.2736874427 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |