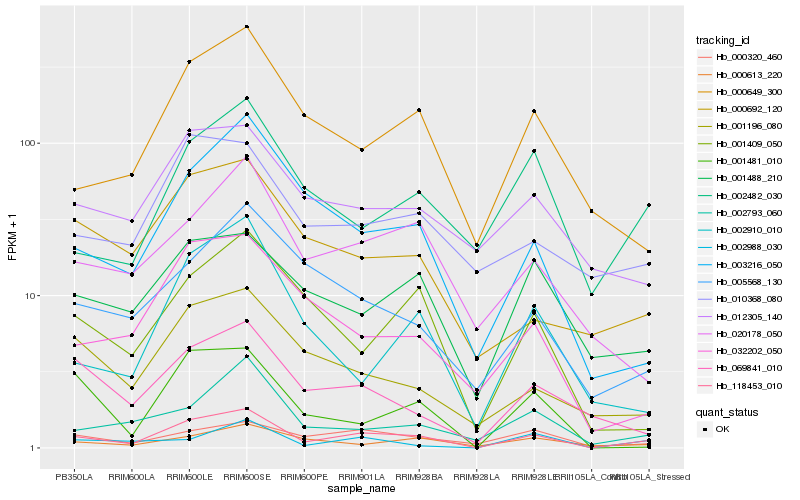
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118453_010 |
0.0 |
- |
- |
hypothetical protein VITISV_012360 [Vitis vinifera] |
| 2 |
Hb_003216_050 |
0.2192498848 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 3 |
Hb_000649_300 |
0.2210867258 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 4 |
Hb_000320_460 |
0.2219277227 |
- |
- |
PREDICTED: uncharacterized protein LOC105650098 [Jatropha curcas] |
| 5 |
Hb_002793_060 |
0.2241163891 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 6 |
Hb_001488_210 |
0.2247887778 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |
| 7 |
Hb_069841_010 |
0.2249787278 |
- |
- |
resistance protein [Quercus suber] |
| 8 |
Hb_001196_080 |
0.2251873644 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 9 |
Hb_010368_080 |
0.2263724855 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 10 |
Hb_032202_050 |
0.2265138912 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_020178_050 |
0.2279407845 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 12 |
Hb_001481_010 |
0.2284840097 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012305_140 |
0.2286003507 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002988_030 |
0.2289974168 |
- |
- |
hypothetical protein EUGRSUZ_K02073 [Eucalyptus grandis] |
| 15 |
Hb_001409_050 |
0.2301916329 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_002482_030 |
0.2302685255 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 17 |
Hb_000692_120 |
0.2317855478 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 18 |
Hb_005568_130 |
0.232165233 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_002910_010 |
0.2328647821 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 20 |
Hb_000613_220 |
0.2335475104 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |