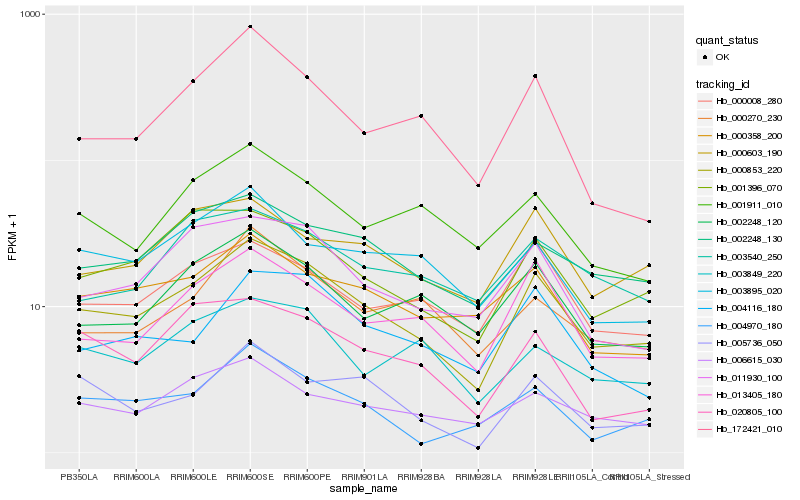
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_220 |
0.0 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 2 |
Hb_013405_180 |
0.112712932 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 3 |
Hb_001396_070 |
0.1161375695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002248_130 |
0.1167128205 |
- |
- |
ubiquitin ligase protein cop1, putative [Ricinus communis] |
| 5 |
Hb_006615_030 |
0.1176472148 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 6 |
Hb_011930_100 |
0.1183524449 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 7 |
Hb_002248_120 |
0.1262547052 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 8 |
Hb_000008_280 |
0.1269589057 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 9 |
Hb_000358_200 |
0.1278444581 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 10 |
Hb_004116_180 |
0.1307080002 |
- |
- |
PREDICTED: uncharacterized protein LOC105646147 [Jatropha curcas] |
| 11 |
Hb_000603_190 |
0.1313726728 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 2-like [Jatropha curcas] |
| 12 |
Hb_172421_010 |
0.1335677275 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 13 |
Hb_003895_020 |
0.1345161334 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 14 |
Hb_003849_220 |
0.1362065766 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 15 |
Hb_001911_010 |
0.1363726499 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 16 |
Hb_000270_230 |
0.1364515137 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |
| 17 |
Hb_005736_050 |
0.1374618495 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 18 |
Hb_003540_250 |
0.1382582489 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |
| 19 |
Hb_020805_100 |
0.138736878 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 20 |
Hb_004970_180 |
0.1389791685 |
- |
- |
- |