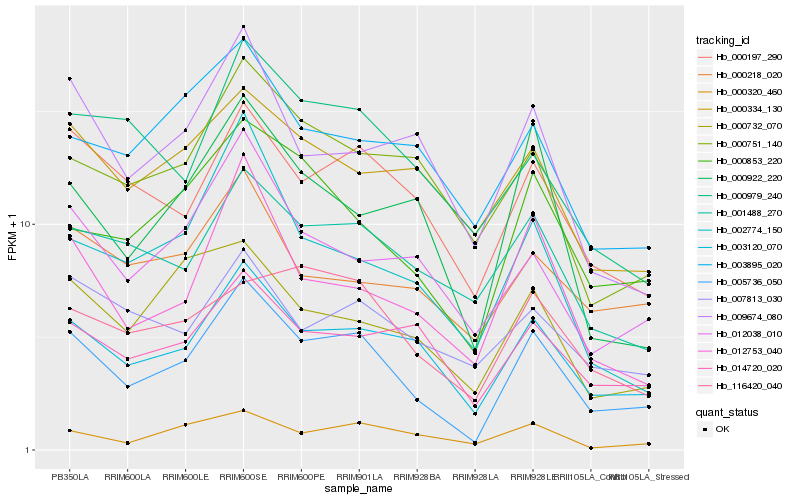
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005736_050 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 2 |
Hb_003120_070 |
0.1149315544 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000853_220 |
0.1374618495 |
- |
- |
PREDICTED: cytidine deaminase 1 [Jatropha curcas] |
| 4 |
Hb_000197_290 |
0.1434040986 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 5 |
Hb_012753_040 |
0.1467623416 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_012038_010 |
0.1574635477 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 7 |
Hb_000922_220 |
0.1585123141 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM-like [Jatropha curcas] |
| 8 |
Hb_001488_270 |
0.1593488261 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 9 |
Hb_000320_460 |
0.1596210393 |
- |
- |
PREDICTED: uncharacterized protein LOC105650098 [Jatropha curcas] |
| 10 |
Hb_014720_020 |
0.1596975842 |
- |
- |
PREDICTED: serine/threonine-protein kinase ATM [Jatropha curcas] |
| 11 |
Hb_000751_140 |
0.1640222606 |
- |
- |
PREDICTED: clathrin interactor EPSIN 2 [Jatropha curcas] |
| 12 |
Hb_002774_150 |
0.1642700613 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 13 |
Hb_009674_080 |
0.1654805441 |
- |
- |
PREDICTED: branchpoint-bridging protein [Jatropha curcas] |
| 14 |
Hb_000732_070 |
0.1663705695 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 15 |
Hb_116420_040 |
0.166894168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000979_240 |
0.1682839389 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein GAIP-B [Jatropha curcas] |
| 17 |
Hb_003895_020 |
0.1718550701 |
- |
- |
PREDICTED: dedicator of cytokinesis protein 7 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000334_130 |
0.1721625241 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 19 |
Hb_000218_020 |
0.1723023377 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 20 |
Hb_007813_030 |
0.1726694649 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |