| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
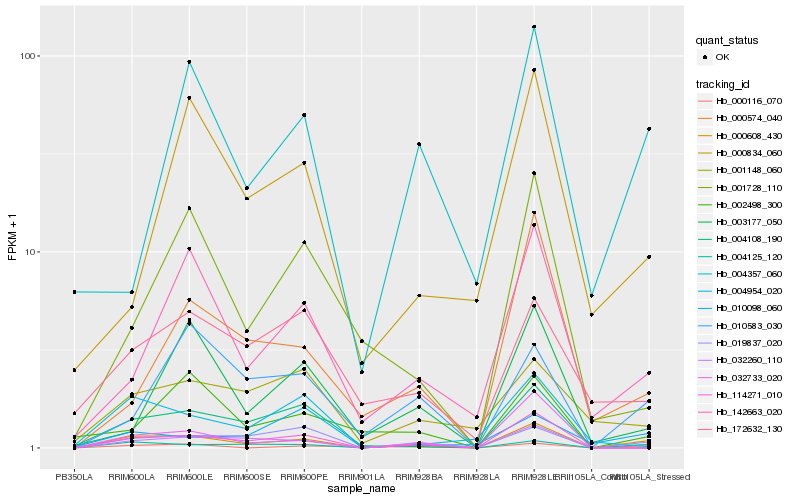
Hb_000608_430 |
0.0 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_004125_120 |
0.1870405924 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_019837_020 |
0.2118714116 |
- |
- |
- |
| 4 |
Hb_114271_010 |
0.2439837728 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 5 |
Hb_142663_020 |
0.2633116361 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 6 |
Hb_032260_110 |
0.2712213763 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000116_070 |
0.2741608641 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 8 |
Hb_010583_030 |
0.284618478 |
- |
- |
hypothetical protein PHAVU_011G114500g, partial [Phaseolus vulgaris] |
| 9 |
Hb_032733_020 |
0.2872872172 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 10 |
Hb_000574_040 |
0.2873177222 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_004357_060 |
0.2916555306 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 12 |
Hb_004108_190 |
0.2932732186 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 13 |
Hb_004954_020 |
0.2964346548 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 14 |
Hb_172632_130 |
0.2978618548 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 15 |
Hb_001148_060 |
0.2980995343 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 16 |
Hb_001728_110 |
0.2983277669 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 17 |
Hb_003177_050 |
0.303814362 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Populus euphratica] |
| 18 |
Hb_002498_300 |
0.3048206926 |
- |
- |
BnaC04g50140D [Brassica napus] |
| 19 |
Hb_000834_060 |
0.3077329198 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_010098_060 |
0.3084427378 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |