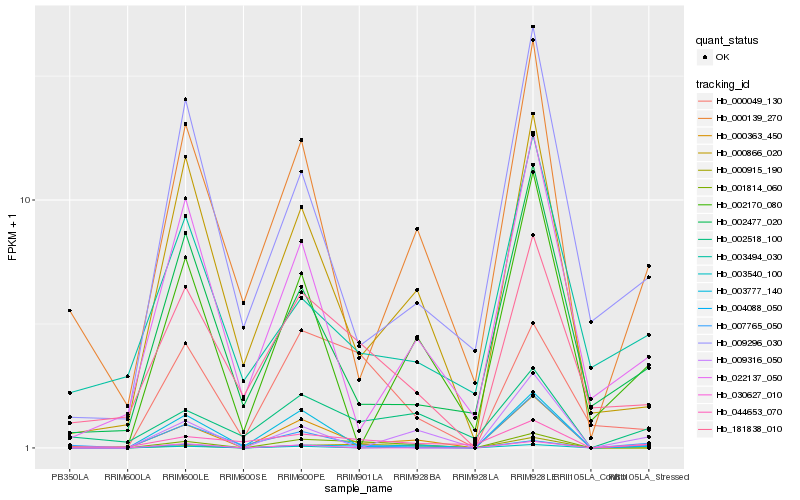
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004088_050 |
0.0 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 2 |
Hb_000363_450 |
0.2184883428 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_000049_130 |
0.249042143 |
- |
- |
PREDICTED: uncharacterized protein LOC104879855 [Vitis vinifera] |
| 4 |
Hb_001814_060 |
0.251952188 |
- |
- |
PREDICTED: uncharacterized protein LOC105650637 [Jatropha curcas] |
| 5 |
Hb_181838_010 |
0.2531865147 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 6 |
Hb_002477_020 |
0.2624846694 |
- |
- |
PREDICTED: uncharacterized protein LOC105631402 [Jatropha curcas] |
| 7 |
Hb_003540_100 |
0.2681832119 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 8 |
Hb_009296_030 |
0.2700517206 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 9 |
Hb_000866_020 |
0.2730207546 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 10 |
Hb_007765_050 |
0.2755526434 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009316_050 |
0.2759490057 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 12 |
Hb_002170_080 |
0.2783621118 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_003777_140 |
0.2799253648 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000139_270 |
0.2808383195 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 15 |
Hb_022137_050 |
0.2817361965 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 16 |
Hb_000915_190 |
0.2855463262 |
- |
- |
- |
| 17 |
Hb_044653_070 |
0.2859463552 |
- |
- |
hypothetical protein CICLE_v10018540mg [Citrus clementina] |
| 18 |
Hb_003494_030 |
0.288593084 |
- |
- |
PREDICTED: protease Do-like 8, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002518_100 |
0.288620787 |
- |
- |
- |
| 20 |
Hb_030627_010 |
0.289521922 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 [Sesamum indicum] |