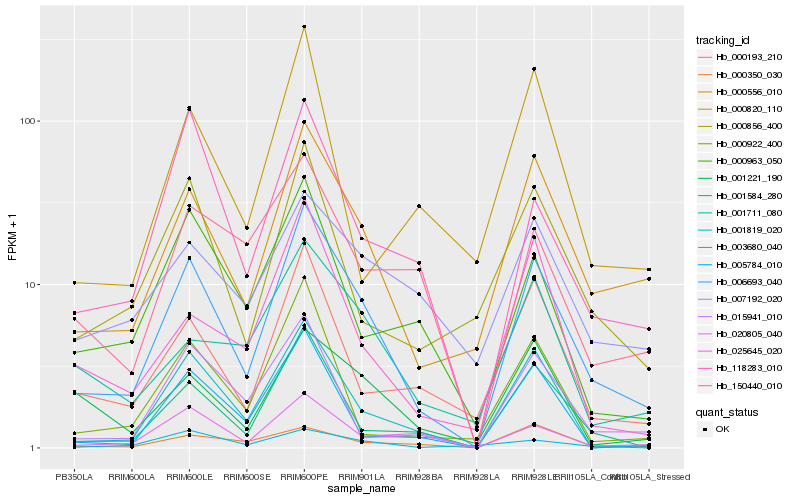
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006693_040 |
0.0 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 2 |
Hb_000556_010 |
0.132511634 |
- |
- |
glycerate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_001819_020 |
0.1580666803 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001584_280 |
0.1651436237 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 5 |
Hb_000193_210 |
0.1883879698 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 6 |
Hb_000856_400 |
0.1905488752 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 7 |
Hb_020805_040 |
0.1908164706 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1A [Jatropha curcas] |
| 8 |
Hb_118283_010 |
0.19980602 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_025645_020 |
0.2016260453 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_015941_010 |
0.2023951517 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_000922_400 |
0.2038155968 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 12 |
Hb_000963_050 |
0.2043390856 |
- |
- |
PREDICTED: vegetative cell wall protein gp1-like [Jatropha curcas] |
| 13 |
Hb_005784_010 |
0.2061366467 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 14 |
Hb_001711_080 |
0.2068630672 |
transcription factor |
TF Family: C2C2-LSD |
PREDICTED: protein LSD1 isoform X1 [Phoenix dactylifera] |
| 15 |
Hb_001221_190 |
0.2107718779 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 16 |
Hb_000350_030 |
0.2115534709 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 17 |
Hb_150440_010 |
0.2180647501 |
- |
- |
- |
| 18 |
Hb_007192_020 |
0.2207790812 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 19 |
Hb_000820_110 |
0.2214271465 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 20 |
Hb_003680_040 |
0.2218016644 |
- |
- |
PREDICTED: polyadenylate-binding protein 7 isoform X1 [Jatropha curcas] |