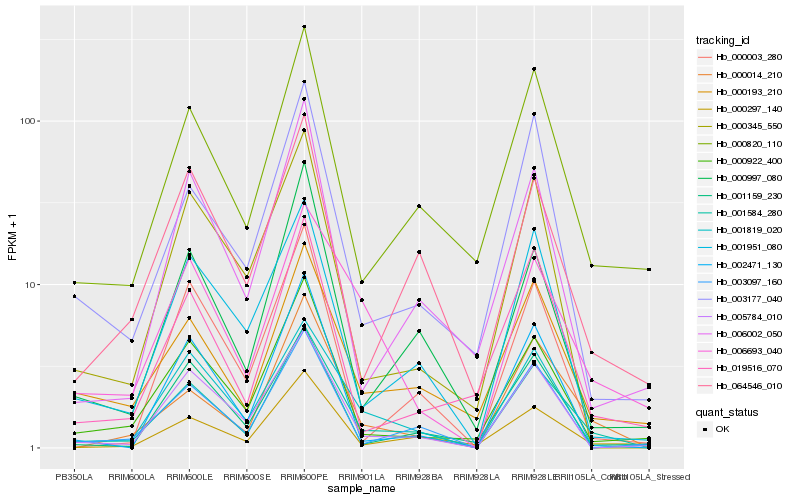
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001584_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 2 |
Hb_000014_210 |
0.1290059042 |
- |
- |
amine oxidase, partial [Aphanizomenon flos-aquae] |
| 3 |
Hb_000820_110 |
0.1414854825 |
- |
- |
UDP-XYL synthase 6 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000193_210 |
0.1433940267 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: galactoside 2-alpha-L-fucosyltransferase [Jatropha curcas] |
| 5 |
Hb_001951_080 |
0.1473410597 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_005784_010 |
0.1511530805 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 7 |
Hb_000297_140 |
0.1540736042 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 8 |
Hb_000922_400 |
0.1547254622 |
- |
- |
PREDICTED: uncharacterized protein LOC105640363 [Jatropha curcas] |
| 9 |
Hb_003097_160 |
0.1549924992 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein sel-10 [Jatropha curcas] |
| 10 |
Hb_003177_040 |
0.1561289148 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 11 |
Hb_000997_080 |
0.1570436274 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001819_020 |
0.1600029521 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002471_130 |
0.1622707108 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 14 |
Hb_064546_010 |
0.1634881809 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 15 |
Hb_006693_040 |
0.1651436237 |
- |
- |
PREDICTED: homeotic protein female sterile-like [Jatropha curcas] |
| 16 |
Hb_019516_070 |
0.1658569975 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 17 |
Hb_006002_050 |
0.1663584415 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 18 |
Hb_000345_550 |
0.1676520456 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 19 |
Hb_001159_230 |
0.1717613063 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 20 |
Hb_000003_280 |
0.1721041219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |