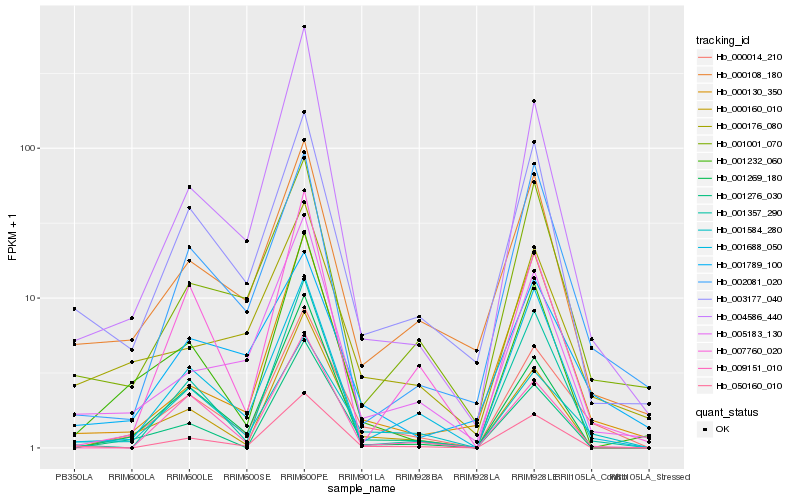
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000014_210 |
0.0 |
- |
- |
amine oxidase, partial [Aphanizomenon flos-aquae] |
| 2 |
Hb_001584_280 |
0.1290059042 |
- |
- |
PREDICTED: uncharacterized protein LOC105647395 [Jatropha curcas] |
| 3 |
Hb_000160_010 |
0.1618252415 |
- |
- |
PREDICTED: uncharacterized protein LOC105638816 [Jatropha curcas] |
| 4 |
Hb_001688_050 |
0.1725284576 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 5 |
Hb_001276_030 |
0.1728257219 |
- |
- |
PREDICTED: uncharacterized protein LOC104881333 [Vitis vinifera] |
| 6 |
Hb_004586_440 |
0.1765877188 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 7 |
Hb_007760_020 |
0.1796057015 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 8 |
Hb_001357_290 |
0.1797787289 |
- |
- |
PREDICTED: uncharacterized protein LOC105632986 [Jatropha curcas] |
| 9 |
Hb_001232_060 |
0.1800517075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001269_180 |
0.1808476452 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_009151_010 |
0.182819796 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 12 |
Hb_003177_040 |
0.1828608426 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 13 |
Hb_001789_100 |
0.1848335452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002081_020 |
0.1851139054 |
transcription factor |
TF Family: LIM |
Cysteine and glycine-rich protein, putative [Ricinus communis] |
| 15 |
Hb_050160_010 |
0.1862413285 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 16 |
Hb_000108_180 |
0.1863452372 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 17 |
Hb_001001_070 |
0.1874414339 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 18 |
Hb_005183_130 |
0.1876895882 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 19 |
Hb_000130_350 |
0.1883332999 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_000176_080 |
0.1888756906 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |