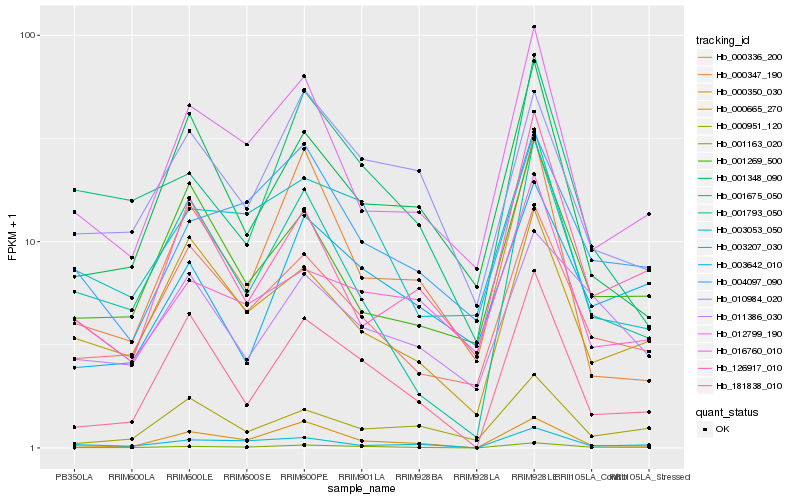
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_020 |
0.0 |
desease resistance |
Gene Name: ABC_trans_N |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_181838_010 |
0.1338439212 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 3 |
Hb_000350_030 |
0.1539598017 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 4 |
Hb_000336_200 |
0.1654295776 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_001793_050 |
0.1687320432 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 6 |
Hb_003207_030 |
0.1713709515 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 7 |
Hb_012799_190 |
0.1747010214 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 8 |
Hb_003642_010 |
0.179692577 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 9 |
Hb_001675_050 |
0.1804624013 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 10 |
Hb_004097_090 |
0.1844983259 |
- |
- |
PREDICTED: uncharacterized protein LOC105634517 [Jatropha curcas] |
| 11 |
Hb_003053_050 |
0.1884955346 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 8 [Jatropha curcas] |
| 12 |
Hb_000665_270 |
0.1904381911 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 13 |
Hb_001348_090 |
0.1953073645 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000951_120 |
0.1961773181 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105636019 [Jatropha curcas] |
| 15 |
Hb_010984_020 |
0.1976585239 |
- |
- |
Translation factor GUF1 like, chloroplastic [Glycine soja] |
| 16 |
Hb_000347_190 |
0.1990925734 |
- |
- |
PREDICTED: transmembrane protein 87A [Jatropha curcas] |
| 17 |
Hb_126917_010 |
0.1991596982 |
- |
- |
PREDICTED: uncharacterized protein LOC105646333 [Jatropha curcas] |
| 18 |
Hb_011386_030 |
0.1994620587 |
- |
- |
PREDICTED: probable protein phosphatase 2C 26 [Jatropha curcas] |
| 19 |
Hb_001269_500 |
0.1997743562 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 20 |
Hb_016760_010 |
0.1999541159 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |