| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
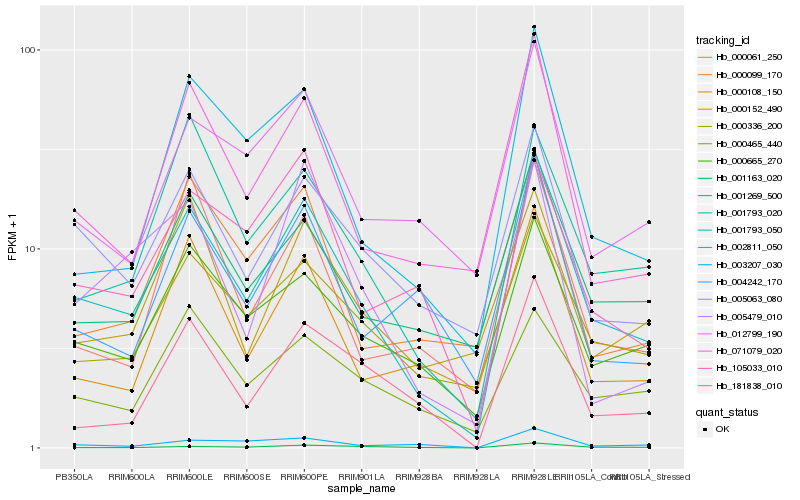
Hb_001793_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 2 |
Hb_000665_270 |
0.1371784638 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_071079_020 |
0.1376395338 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 4 |
Hb_000336_200 |
0.1398849105 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_005063_080 |
0.1405077243 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002811_050 |
0.142143193 |
- |
- |
PREDICTED: uncharacterized protein LOC105647652 [Jatropha curcas] |
| 7 |
Hb_001269_500 |
0.1471874672 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 8 |
Hb_001793_020 |
0.1543801813 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 9 |
Hb_012799_190 |
0.1566692446 |
- |
- |
PREDICTED: uncharacterized protein LOC105648490 [Jatropha curcas] |
| 10 |
Hb_000108_150 |
0.163083711 |
- |
- |
alpha/beta hydrolase, putative [Ricinus communis] |
| 11 |
Hb_000099_170 |
0.1650139476 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 12 |
Hb_181838_010 |
0.1656290322 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 13 |
Hb_000061_250 |
0.1663980529 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 14 |
Hb_105033_010 |
0.1678630986 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004242_170 |
0.167982467 |
- |
- |
calcium/calmodulin-regulated receptor-like kinase [Manihot esculenta] |
| 16 |
Hb_001163_020 |
0.1687320432 |
desease resistance |
Gene Name: ABC_trans_N |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_000465_440 |
0.1694648672 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 18 |
Hb_000152_490 |
0.1713874254 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 19 |
Hb_005479_010 |
0.1736231453 |
- |
- |
PREDICTED: uncharacterized protein LOC105111002 [Populus euphratica] |
| 20 |
Hb_003207_030 |
0.1739543789 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |