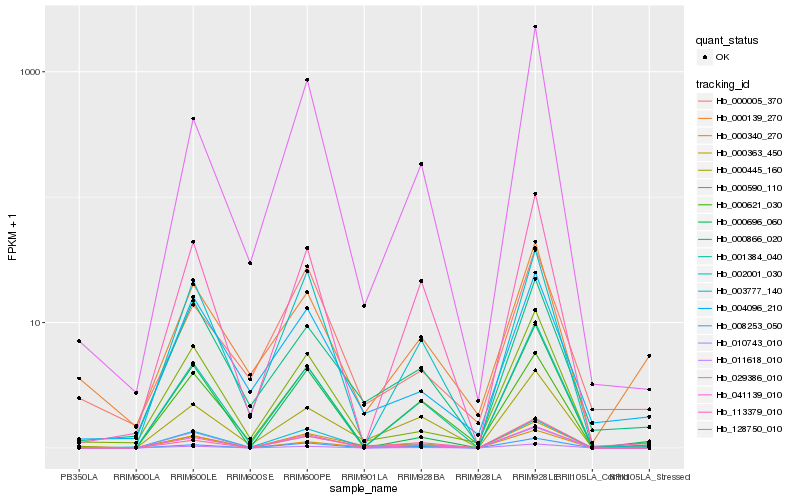
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_450 |
0.0 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 1 [Jatropha curcas] |
| 2 |
Hb_003777_140 |
0.1612822925 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_128750_010 |
0.1663499574 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 4 |
Hb_010743_010 |
0.1704232652 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000445_160 |
0.1794165396 |
- |
- |
PREDICTED: probable calcium-binding protein CML44 [Jatropha curcas] |
| 6 |
Hb_000866_020 |
0.1823085603 |
- |
- |
hypothetical protein B456_004G042000 [Gossypium raimondii] |
| 7 |
Hb_041139_010 |
0.1827051563 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 8 |
Hb_000340_270 |
0.1833077086 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 9 |
Hb_008253_050 |
0.1834745495 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 10 |
Hb_011618_010 |
0.1850563546 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 11 |
Hb_000590_110 |
0.1872963501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001384_040 |
0.1883587837 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 13 |
Hb_000139_270 |
0.1913091547 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 14 |
Hb_000621_030 |
0.1931594199 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 15 |
Hb_113379_010 |
0.1944440457 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 16 |
Hb_000005_370 |
0.194591624 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105634024 [Jatropha curcas] |
| 17 |
Hb_000696_060 |
0.1946236238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004096_210 |
0.1947514987 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 19 |
Hb_029386_010 |
0.1953799491 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 20 |
Hb_002001_030 |
0.1963527579 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |