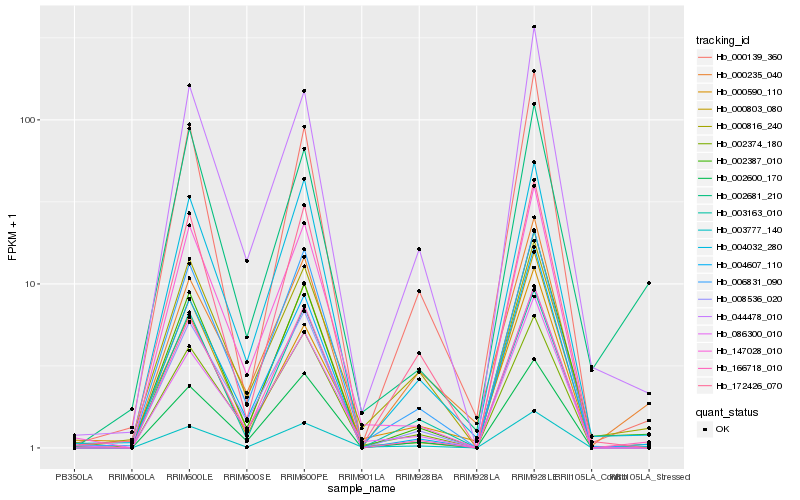
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003777_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_166718_010 |
0.1040685601 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 3 |
Hb_006831_090 |
0.1153352569 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002387_010 |
0.1164033336 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 5 |
Hb_086300_010 |
0.1170010308 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_172426_070 |
0.1180863744 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000139_360 |
0.1186843363 |
- |
- |
PREDICTED: uncharacterized protein LOC105631933 [Jatropha curcas] |
| 8 |
Hb_004032_280 |
0.1188741187 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 9 |
Hb_003163_010 |
0.1210425095 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 10 |
Hb_000803_080 |
0.1224200972 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 11 |
Hb_000816_240 |
0.1235543775 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_147028_010 |
0.1236963576 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 13 |
Hb_004607_110 |
0.1243216753 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 14 |
Hb_002374_180 |
0.1244454746 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 15 |
Hb_008536_020 |
0.1246133151 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 16 |
Hb_002681_210 |
0.1261378006 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 17 |
Hb_000235_040 |
0.1263682277 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 18 |
Hb_002600_170 |
0.1268778234 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_044478_010 |
0.1271278594 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 20 |
Hb_000590_110 |
0.1276023334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |