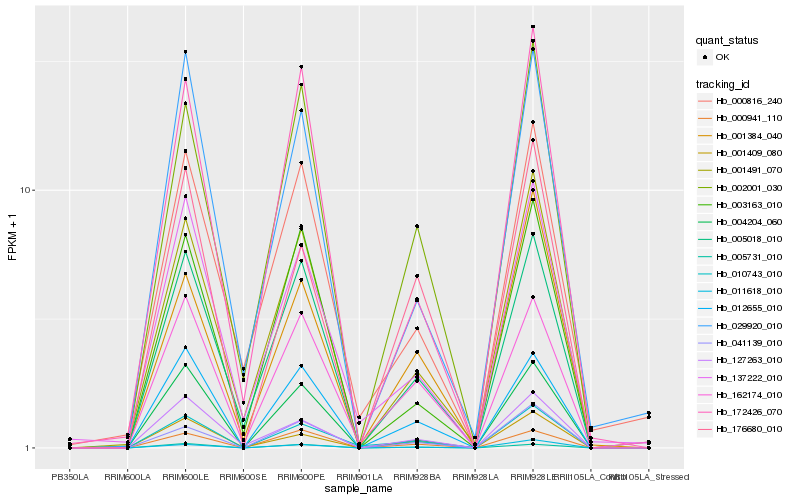
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010743_010 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_137222_010 |
0.1148649494 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 3 |
Hb_012655_010 |
0.1254088674 |
- |
- |
hypothetical protein JCGZ_09061 [Jatropha curcas] |
| 4 |
Hb_004204_060 |
0.1269423043 |
transcription factor |
TF Family: NAC |
NAC transcription factor 058, partial [Jatropha curcas] |
| 5 |
Hb_001409_080 |
0.1316955452 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Populus euphratica] |
| 6 |
Hb_001491_070 |
0.1323403144 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 7 |
Hb_162174_010 |
0.1324854342 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_011618_010 |
0.1336971801 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 9 |
Hb_005018_010 |
0.135312169 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_041139_010 |
0.1363088681 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 11 |
Hb_127263_010 |
0.1372727631 |
- |
- |
hypothetical protein JCGZ_02828 [Jatropha curcas] |
| 12 |
Hb_002001_030 |
0.1375847591 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 13 |
Hb_176680_010 |
0.1385009915 |
- |
- |
hypothetical protein JCGZ_22787 [Jatropha curcas] |
| 14 |
Hb_172426_070 |
0.1392554539 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001384_040 |
0.1421941023 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 16 |
Hb_005731_010 |
0.1441450134 |
- |
- |
hypothetical protein VITISV_030941 [Vitis vinifera] |
| 17 |
Hb_003163_010 |
0.1471062155 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 18 |
Hb_029920_010 |
0.1484101422 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 19 |
Hb_000816_240 |
0.1490158557 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000941_110 |
0.149829443 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |