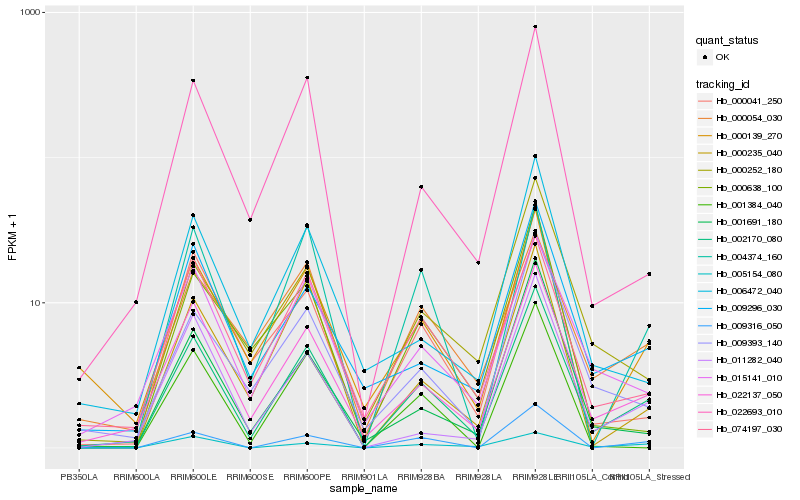
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002170_080 |
0.0 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 2 |
Hb_022137_050 |
0.1163547086 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 1-like [Jatropha curcas] |
| 3 |
Hb_009316_050 |
0.1425033825 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 4 |
Hb_022693_010 |
0.1551502249 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 5 |
Hb_000041_250 |
0.1570691695 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 6 |
Hb_000235_040 |
0.1608413139 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X1 [Populus euphratica] |
| 7 |
Hb_009393_140 |
0.1647948528 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 8 |
Hb_009296_030 |
0.165195008 |
- |
- |
PREDICTED: uncharacterized protein LOC105634328 [Jatropha curcas] |
| 9 |
Hb_000252_180 |
0.1662679911 |
- |
- |
Glutamate--glyoxylate aminotransferase 2 [Morus notabilis] |
| 10 |
Hb_015141_010 |
0.1663301699 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 11 |
Hb_074197_030 |
0.1679652535 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_004374_160 |
0.1683004499 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 13 |
Hb_011282_040 |
0.1692089768 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 14 |
Hb_000139_270 |
0.1721845585 |
- |
- |
hypothetical protein POPTR_0008s14890g [Populus trichocarpa] |
| 15 |
Hb_001691_180 |
0.1749561835 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 16 |
Hb_000638_100 |
0.1769533405 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 17 |
Hb_005154_080 |
0.1776787703 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105640764 [Jatropha curcas] |
| 18 |
Hb_001384_040 |
0.1802721585 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 19 |
Hb_000054_030 |
0.1813415585 |
- |
- |
hypothetical protein POPTR_0010s17680g [Populus trichocarpa] |
| 20 |
Hb_006472_040 |
0.181457763 |
- |
- |
PREDICTED: inner membrane protein PPF-1, chloroplastic [Jatropha curcas] |