| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
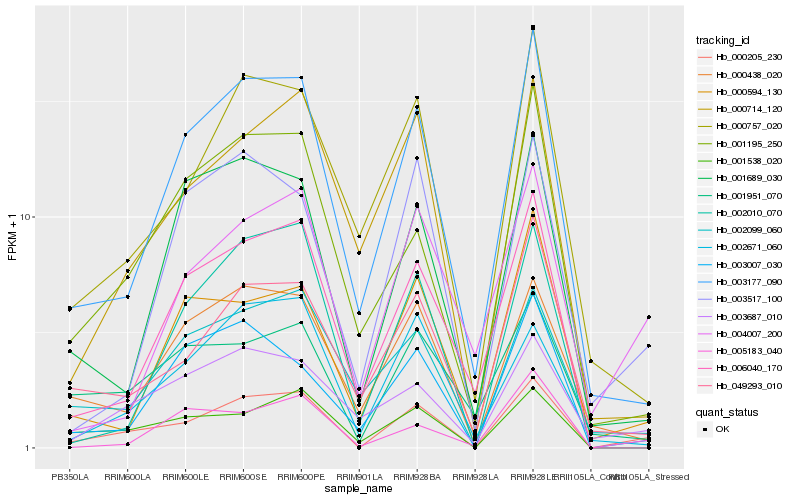
Hb_000205_230 |
0.0 |
transcription factor |
TF Family: zf-HD |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_003177_090 |
0.1710258106 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 3 |
Hb_049293_010 |
0.1913370838 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_001195_250 |
0.1913883037 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_001538_020 |
0.1956824061 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 6 |
Hb_002099_060 |
0.196253993 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 7 |
Hb_000757_020 |
0.1968532229 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_000714_120 |
0.1969192362 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 9 |
Hb_002671_060 |
0.1987193448 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 10 |
Hb_003687_010 |
0.1988413659 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 11 |
Hb_000438_020 |
0.1992163523 |
- |
- |
leucine-rich repeat-containing protein 2, lrrc2, putative [Ricinus communis] |
| 12 |
Hb_001689_030 |
0.2017017468 |
- |
- |
PREDICTED: J domain-containing protein required for chloroplast accumulation response 1 [Jatropha curcas] |
| 13 |
Hb_001951_070 |
0.2039415352 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 14 |
Hb_003517_100 |
0.2043976015 |
transcription factor |
TF Family: GRAS |
- |
| 15 |
Hb_002010_070 |
0.2050829993 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 16 |
Hb_003007_030 |
0.2056293866 |
- |
- |
- |
| 17 |
Hb_006040_170 |
0.2067949334 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 18 |
Hb_004007_200 |
0.2070614385 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein NPY5 [Jatropha curcas] |
| 19 |
Hb_005183_040 |
0.2071127905 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 20 |
Hb_000594_130 |
0.2076317346 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |