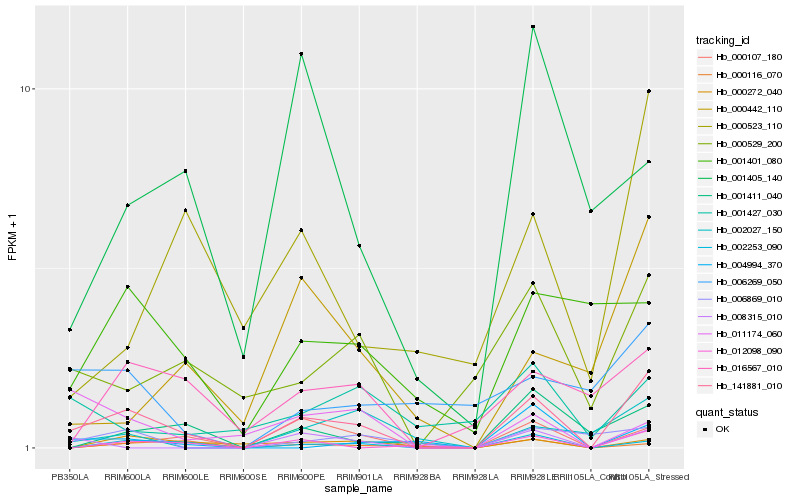
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141881_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_002253_090 |
0.2859363878 |
- |
- |
Chitinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_012098_090 |
0.3230749426 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_000107_180 |
0.3253988893 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 5 |
Hb_000529_200 |
0.3292673354 |
- |
- |
- |
| 6 |
Hb_016567_010 |
0.3545845484 |
- |
- |
- |
| 7 |
Hb_000272_040 |
0.35479301 |
- |
- |
- |
| 8 |
Hb_001411_040 |
0.3552037888 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 9 |
Hb_008315_010 |
0.3600551485 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 10 |
Hb_000523_110 |
0.363360009 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 11 |
Hb_001401_080 |
0.3653549054 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006869_010 |
0.3657995189 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_011174_060 |
0.3670846815 |
- |
- |
- |
| 14 |
Hb_002027_150 |
0.367096926 |
- |
- |
- |
| 15 |
Hb_000442_110 |
0.3678884342 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 16 |
Hb_000116_070 |
0.3681274935 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22381 [Jatropha curcas] |
| 17 |
Hb_001405_140 |
0.3734891831 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 18 |
Hb_004994_370 |
0.3762100076 |
- |
- |
- |
| 19 |
Hb_001427_030 |
0.3823515784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006269_050 |
0.3848275273 |
- |
- |
hypothetical protein POPTR_0004s13450g [Populus trichocarpa] |