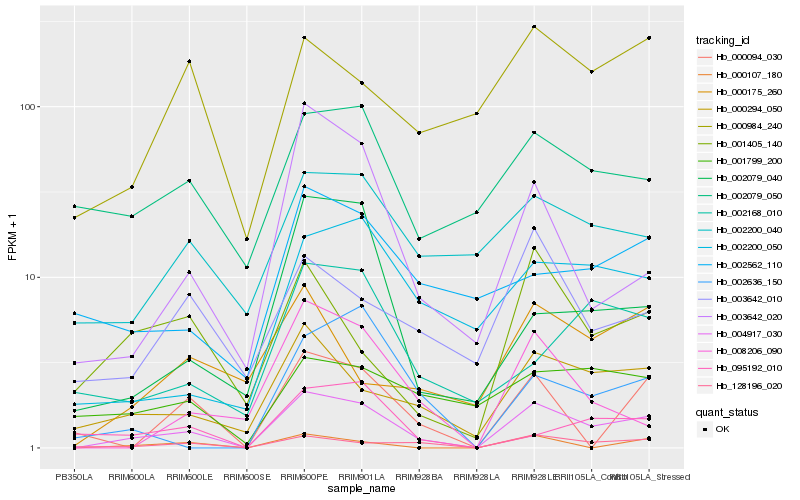
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004917_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_128196_020 |
0.2135358332 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Nelumbo nucifera] |
| 3 |
Hb_000294_050 |
0.2223733611 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 4 |
Hb_003642_020 |
0.2624848129 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 5 |
Hb_001405_140 |
0.2690767211 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 6 |
Hb_002200_040 |
0.2695944495 |
- |
- |
PREDICTED: uncharacterized protein LOC105629143 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002200_050 |
0.2708766891 |
- |
- |
unknown [Lotus japonicus] |
| 8 |
Hb_000107_180 |
0.2715222571 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103343662 [Prunus mume] |
| 9 |
Hb_002079_040 |
0.2746750547 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |
| 10 |
Hb_008206_090 |
0.2752748998 |
- |
- |
PREDICTED: uncharacterized protein LOC100811033 [Glycine max] |
| 11 |
Hb_003642_010 |
0.2766535504 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_002636_150 |
0.2769374395 |
- |
- |
- |
| 13 |
Hb_002079_050 |
0.2769426812 |
- |
- |
hypothetical protein B456_004G182300 [Gossypium raimondii] |
| 14 |
Hb_000094_030 |
0.2770745005 |
- |
- |
- |
| 15 |
Hb_095192_010 |
0.2773521414 |
- |
- |
hypothetical protein VITISV_043980 [Vitis vinifera] |
| 16 |
Hb_002168_010 |
0.2809433467 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 17 |
Hb_000984_240 |
0.2812853576 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 18 |
Hb_001799_200 |
0.2831514624 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 19 |
Hb_000175_260 |
0.2835702214 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 20 |
Hb_002562_110 |
0.2893531079 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |