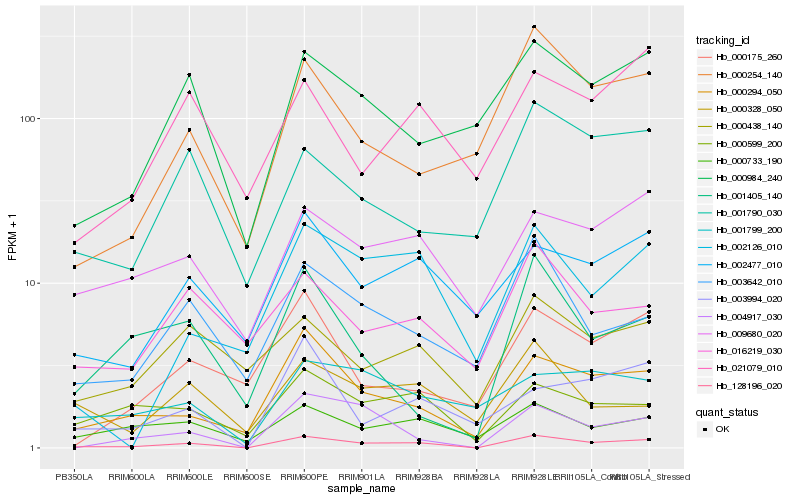
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128196_020 |
0.0 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 3, chloroplastic [Nelumbo nucifera] |
| 2 |
Hb_000294_050 |
0.1846957949 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 3 |
Hb_004917_030 |
0.2135358332 |
- |
- |
- |
| 4 |
Hb_003642_010 |
0.2147983835 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_009680_020 |
0.2154850693 |
- |
- |
PREDICTED: 40S ribosomal protein S10 [Jatropha curcas] |
| 6 |
Hb_000599_200 |
0.2167428528 |
- |
- |
ABC transporter family protein [Populus trichocarpa] |
| 7 |
Hb_000733_190 |
0.2188064257 |
- |
- |
zinc ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_000984_240 |
0.2188402256 |
- |
- |
hypothetical protein JCGZ_15962 [Jatropha curcas] |
| 9 |
Hb_000254_140 |
0.2190321648 |
- |
- |
PREDICTED: 30S ribosomal protein S17, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002477_010 |
0.2198746528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002126_010 |
0.2271682352 |
- |
- |
predicted protein [Hordeum vulgare subsp. vulgare] |
| 12 |
Hb_001790_030 |
0.2283478071 |
- |
- |
PREDICTED: ferredoxin-thioredoxin reductase catalytic chain, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000438_140 |
0.2289735422 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 14 |
Hb_016219_030 |
0.2339383575 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_003994_020 |
0.2346299002 |
- |
- |
- |
| 16 |
Hb_000328_050 |
0.2375496428 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001405_140 |
0.2378271436 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 18 |
Hb_021079_010 |
0.239514949 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 19 |
Hb_001799_200 |
0.2429309106 |
- |
- |
PREDICTED: tRNA wybutosine-synthesizing protein 4 [Jatropha curcas] |
| 20 |
Hb_000175_260 |
0.2436578458 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |