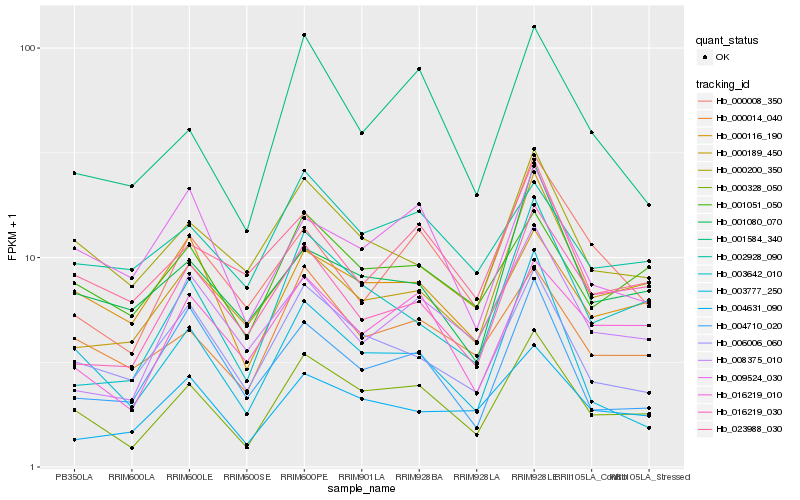
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000328_050 |
0.0 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000014_040 |
0.1234343635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003642_010 |
0.1245958935 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_001584_340 |
0.1257363679 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 5 |
Hb_006006_060 |
0.1267345807 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_016219_010 |
0.1271757333 |
- |
- |
hypothetical protein B456_001G213800 [Gossypium raimondii] |
| 7 |
Hb_003777_250 |
0.1352738715 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 8 |
Hb_001051_050 |
0.1371369652 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000200_350 |
0.1390600967 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 10 |
Hb_004631_090 |
0.1446671797 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 11 |
Hb_016219_030 |
0.1465702533 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_001080_070 |
0.1472055312 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 13 |
Hb_000116_190 |
0.1472718986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000189_450 |
0.1496927385 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_008375_010 |
0.1517036339 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 16 |
Hb_023988_030 |
0.1524232729 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 17 |
Hb_000008_350 |
0.1532384318 |
- |
- |
- |
| 18 |
Hb_004710_020 |
0.1535704214 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_009524_030 |
0.1542513071 |
- |
- |
PREDICTED: uncharacterized protein LOC105650971 [Jatropha curcas] |
| 20 |
Hb_002928_090 |
0.154581761 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |