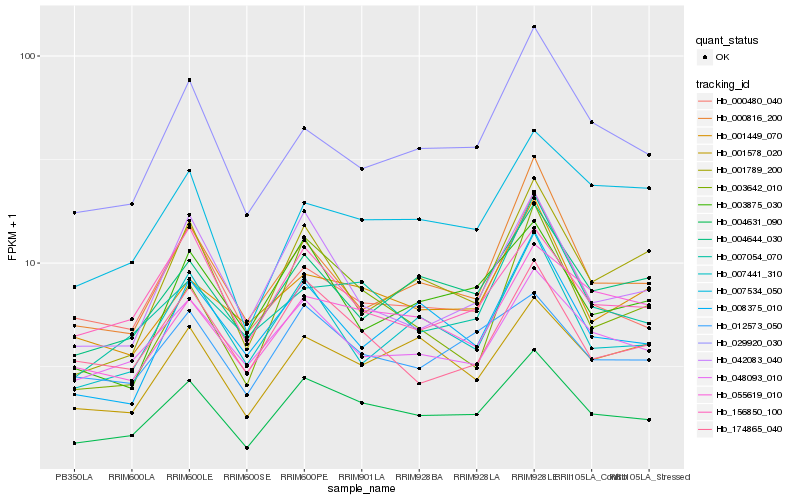
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004631_090 |
0.0 |
- |
- |
PREDICTED: probable glucan 1,3-beta-glucosidase A [Jatropha curcas] |
| 2 |
Hb_012573_050 |
0.1008782823 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 3 |
Hb_004644_030 |
0.1061712514 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 4 |
Hb_048093_010 |
0.1064000195 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_007441_310 |
0.1081856436 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_156850_100 |
0.1095215147 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 7 |
Hb_042083_040 |
0.1099715869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_174865_040 |
0.1107582243 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_029920_030 |
0.1120147862 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001789_200 |
0.1133718868 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001578_020 |
0.113612306 |
- |
- |
Protein virR, putative [Ricinus communis] |
| 12 |
Hb_000480_040 |
0.1137186452 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 13 |
Hb_003875_030 |
0.1141666346 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 14 |
Hb_008375_010 |
0.1143564931 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 15 |
Hb_000816_200 |
0.1177404817 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 16 |
Hb_003642_010 |
0.1191199707 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 17 |
Hb_007054_070 |
0.1203997396 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 18 |
Hb_055619_010 |
0.1212299671 |
- |
- |
PREDICTED: uncharacterized protein LOC105632456 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007534_050 |
0.1218200006 |
- |
- |
PREDICTED: glutamyl-tRNA reductase-binding protein, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001449_070 |
0.1222472221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |