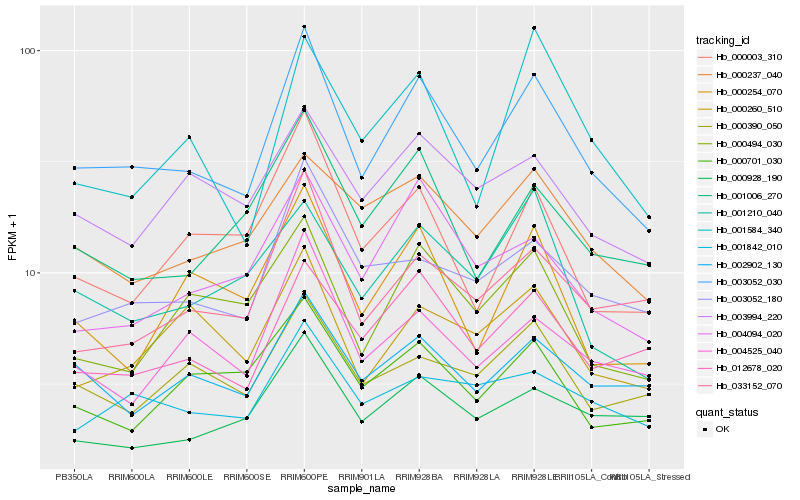
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003052_030 |
0.0 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 2 |
Hb_012678_020 |
0.1127853584 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001006_270 |
0.11758177 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 4 |
Hb_004525_040 |
0.1200627623 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 5 |
Hb_002902_130 |
0.1203047293 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 6 |
Hb_033152_070 |
0.120592531 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 7 |
Hb_003052_180 |
0.1209922 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 8 |
Hb_000494_030 |
0.122656512 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 9 |
Hb_004094_020 |
0.123881054 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000003_310 |
0.1245802408 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 11 |
Hb_003994_220 |
0.124667496 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 12 |
Hb_000237_040 |
0.1248846245 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 13 |
Hb_000254_070 |
0.1252177459 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 14 |
Hb_001842_010 |
0.1254948363 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000260_510 |
0.1258444348 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000390_050 |
0.1265792245 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 17 |
Hb_000701_030 |
0.1271124372 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 18 |
Hb_001210_040 |
0.1279133562 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 19 |
Hb_001584_340 |
0.1279158481 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 20 |
Hb_000928_190 |
0.1297618551 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |