| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
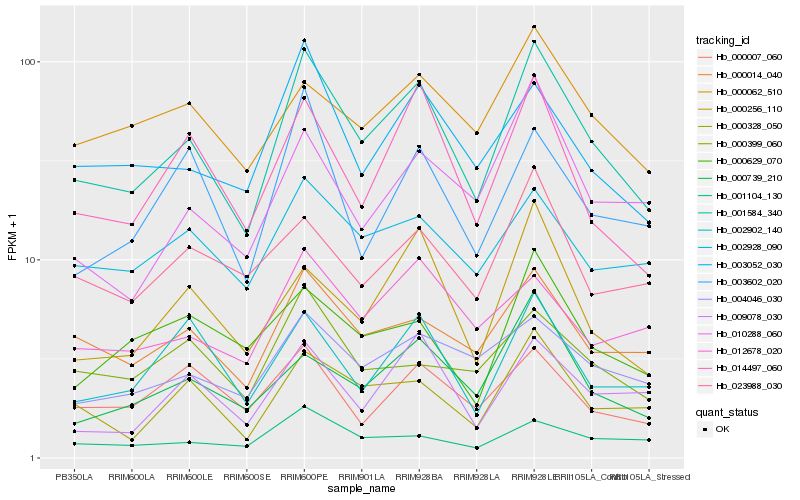
Hb_001584_340 |
0.0 |
- |
- |
PREDICTED: ATP-citrate synthase alpha chain protein 2 [Jatropha curcas] |
| 2 |
Hb_000014_040 |
0.1201020433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000328_050 |
0.1257363679 |
- |
- |
PREDICTED: translation initiation factor IF-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_014497_060 |
0.127792998 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 5 |
Hb_003052_030 |
0.1279158481 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_004046_030 |
0.1375561975 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 7 |
Hb_000739_210 |
0.1395966364 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 8 |
Hb_000256_110 |
0.1396106904 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001104_130 |
0.1415886826 |
- |
- |
C-14 sterol reductase, putative [Ricinus communis] |
| 10 |
Hb_002928_090 |
0.1416679551 |
- |
- |
PREDICTED: metallocarboxypeptidase A-like protein TRV_02598 [Jatropha curcas] |
| 11 |
Hb_002902_140 |
0.1431395856 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 9 [Jatropha curcas] |
| 12 |
Hb_000062_510 |
0.144517483 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_010288_060 |
0.1472248909 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 14 |
Hb_003602_020 |
0.1478773012 |
- |
- |
PREDICTED: serine hydroxymethyltransferase, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_009078_030 |
0.1484510686 |
- |
- |
hypothetical protein PRUPE_ppa016992mg, partial [Prunus persica] |
| 16 |
Hb_012678_020 |
0.1497555593 |
- |
- |
PREDICTED: outer envelope protein 61 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000399_060 |
0.1504824383 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 18 |
Hb_023988_030 |
0.1519258236 |
- |
- |
PREDICTED: transmembrane protein 19 [Vitis vinifera] |
| 19 |
Hb_000007_060 |
0.152202106 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000629_070 |
0.1528274058 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |