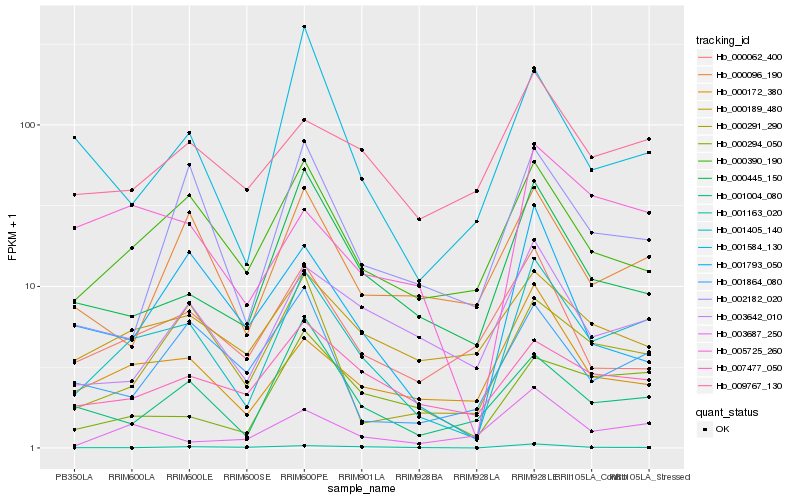
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001405_140 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s08350g, partial [Populus trichocarpa] |
| 2 |
Hb_000390_190 |
0.1865467719 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000172_380 |
0.1958912362 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 4 |
Hb_000445_150 |
0.2003171315 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 5 |
Hb_003642_010 |
0.2075274781 |
- |
- |
PREDICTED: chlorophyll(ide) b reductase NOL, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_000291_290 |
0.2094153857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007477_050 |
0.2098959804 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001004_080 |
0.213547001 |
- |
- |
hypothetical protein POPTR_0014s14130g [Populus trichocarpa] |
| 9 |
Hb_000294_050 |
0.213582357 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 10 |
Hb_009767_130 |
0.2140146657 |
- |
- |
PREDICTED: preprotein translocase subunit SECE1 [Jatropha curcas] |
| 11 |
Hb_005725_260 |
0.2142808613 |
- |
- |
squalene monooxygenase [Jatropha curcas] |
| 12 |
Hb_002182_020 |
0.2149095279 |
- |
- |
Thylakoid lumenal 17.4 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000096_190 |
0.216142096 |
- |
- |
unknown [Lotus japonicus] |
| 14 |
Hb_001163_020 |
0.2175150052 |
desease resistance |
Gene Name: ABC_trans_N |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_001864_080 |
0.2176003284 |
- |
- |
PREDICTED: uncharacterized protein LOC105649405 [Jatropha curcas] |
| 16 |
Hb_000189_480 |
0.2194519531 |
- |
- |
PREDICTED: uncharacterized protein LOC105111904 isoform X2 [Populus euphratica] |
| 17 |
Hb_001793_050 |
0.2231310589 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 18 |
Hb_003687_250 |
0.2234337931 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 19 |
Hb_000062_400 |
0.2234778811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001584_130 |
0.2246660937 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |