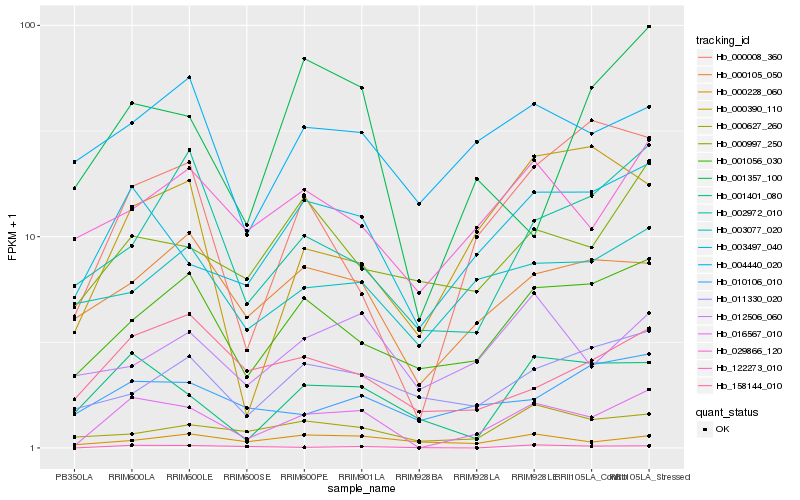
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016567_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_003497_040 |
0.1998201238 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001056_030 |
0.2223087252 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 16 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001401_080 |
0.2311892472 |
- |
- |
PREDICTED: protein TIC 22, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000008_360 |
0.2335722023 |
- |
- |
hypothetical protein POPTR_0004s22050g [Populus trichocarpa] |
| 6 |
Hb_000105_050 |
0.2380993696 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 7 |
Hb_029866_120 |
0.2446254858 |
- |
- |
hypothetical protein CICLE_v10006070mg [Citrus clementina] |
| 8 |
Hb_002972_010 |
0.2517030367 |
- |
- |
PREDICTED: magnesium-dependent phosphatase 1 [Jatropha curcas] |
| 9 |
Hb_158144_010 |
0.2531786297 |
- |
- |
- |
| 10 |
Hb_004440_020 |
0.2538601615 |
- |
- |
AP-3 complex subunit sigma-1, putative [Ricinus communis] |
| 11 |
Hb_000997_250 |
0.2541385785 |
- |
- |
PREDICTED: probable alpha-mannosidase I MNS4 [Jatropha curcas] |
| 12 |
Hb_000627_260 |
0.2545247183 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 13 |
Hb_000390_110 |
0.2556744373 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 14 |
Hb_011330_020 |
0.2573079087 |
transcription factor |
TF Family: ARID |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_012506_060 |
0.2581154568 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 16 |
Hb_122273_010 |
0.2584119854 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_000228_060 |
0.2593609286 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 18 |
Hb_003077_020 |
0.259718824 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001357_100 |
0.2604426351 |
- |
- |
PREDICTED: small ubiquitin-related modifier 1 [Jatropha curcas] |
| 20 |
Hb_010106_010 |
0.2618724569 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |