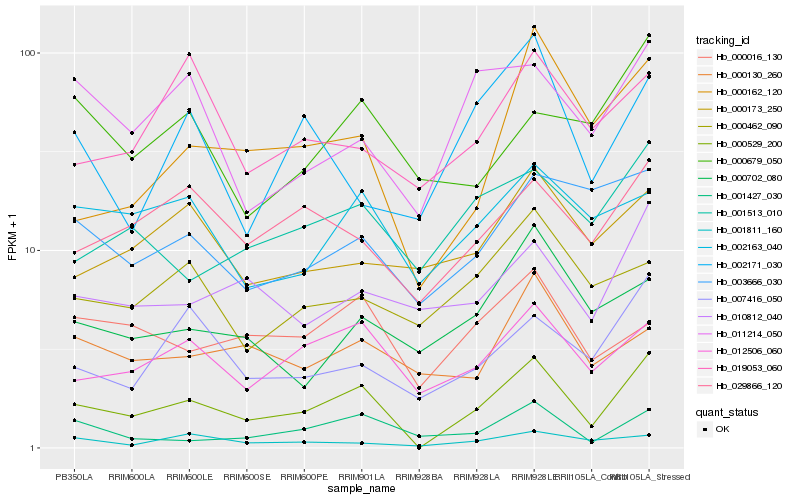
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_200 |
0.0 |
- |
- |
- |
| 2 |
Hb_012506_060 |
0.1943091367 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 3 |
Hb_000162_120 |
0.2017811431 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_007416_050 |
0.2096623461 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 5 [Jatropha curcas] |
| 5 |
Hb_029866_120 |
0.2117619938 |
- |
- |
hypothetical protein CICLE_v10006070mg [Citrus clementina] |
| 6 |
Hb_001427_030 |
0.2130171382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010812_040 |
0.2203248064 |
transcription factor |
TF Family: SNF2 |
PREDICTED: E3 ubiquitin-protein ligase SHPRH [Jatropha curcas] |
| 8 |
Hb_001513_010 |
0.2231798419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011214_050 |
0.2238415931 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 10 |
Hb_000016_130 |
0.2272653952 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase [Jatropha curcas] |
| 11 |
Hb_001811_160 |
0.2276325354 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Eucalyptus grandis] |
| 12 |
Hb_003666_030 |
0.2276682957 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 13 |
Hb_002163_040 |
0.2280658564 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000130_260 |
0.2349930914 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000679_050 |
0.236070821 |
- |
- |
Alba DNA/RNA-binding protein [Theobroma cacao] |
| 16 |
Hb_000702_080 |
0.2371831061 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |
| 17 |
Hb_000173_250 |
0.2373978101 |
- |
- |
PREDICTED: ribonucleoside-diphosphate reductase small chain A [Jatropha curcas] |
| 18 |
Hb_019053_060 |
0.2377034357 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 19 |
Hb_002171_030 |
0.2394812353 |
- |
- |
plastid acyl-ACP thioesterase [Vernicia fordii] |
| 20 |
Hb_000462_090 |
0.2395869754 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |