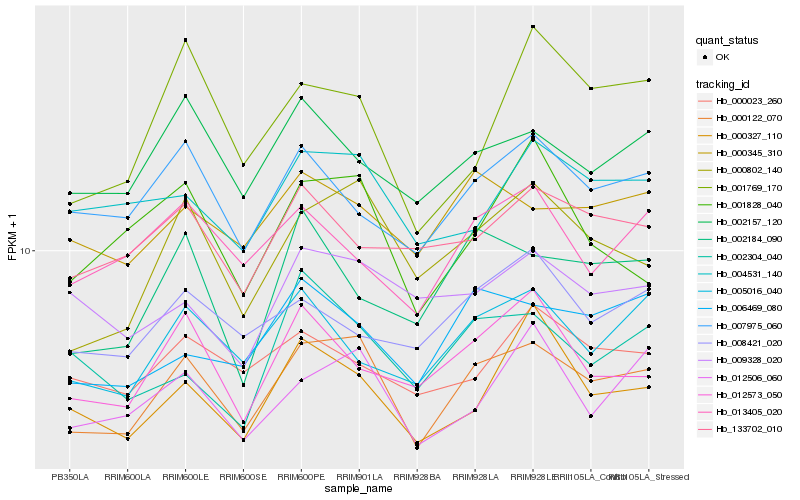
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000122_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 2 |
Hb_000802_140 |
0.0894382892 |
- |
- |
hypothetical protein B456_011G045700 [Gossypium raimondii] |
| 3 |
Hb_012506_060 |
0.1222527975 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 4 |
Hb_006469_080 |
0.123516008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002157_120 |
0.1274653037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000345_310 |
0.1284534036 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein isoform X3 [Jatropha curcas] |
| 7 |
Hb_004531_140 |
0.1299290917 |
- |
- |
PREDICTED: uncharacterized protein LOC105635023 [Jatropha curcas] |
| 8 |
Hb_007975_060 |
0.1303995031 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005016_040 |
0.1316972282 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 10 |
Hb_001828_040 |
0.1328302841 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 11 |
Hb_001769_170 |
0.1335207126 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 12 |
Hb_002184_090 |
0.1341308895 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_012573_050 |
0.1343668834 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 14 |
Hb_000327_110 |
0.1357273123 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 15 |
Hb_013405_020 |
0.1372308106 |
- |
- |
- |
| 16 |
Hb_009328_020 |
0.13862516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000023_260 |
0.1401504451 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002304_040 |
0.1415114097 |
- |
- |
PREDICTED: uncharacterized protein LOC105649623 isoform X2 [Jatropha curcas] |
| 19 |
Hb_133702_010 |
0.1415500971 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 20 |
Hb_008421_020 |
0.1423157073 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |