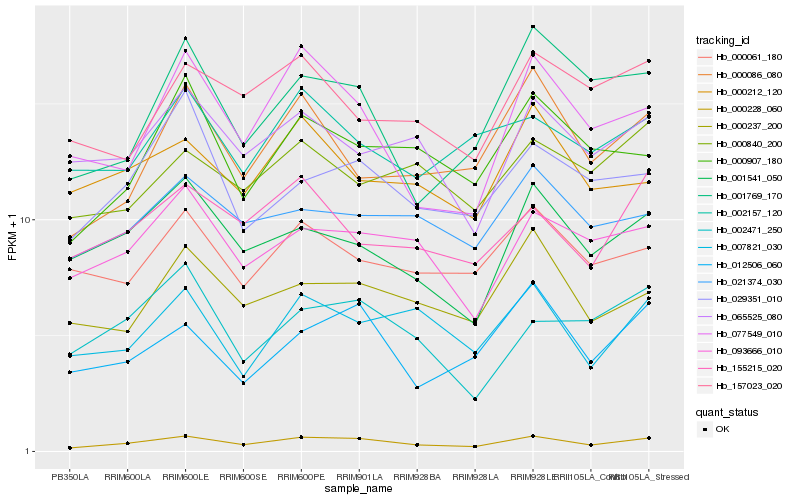
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000228_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_21532 [Jatropha curcas] |
| 2 |
Hb_093666_010 |
0.1107374601 |
- |
- |
BnaC05g06620D [Brassica napus] |
| 3 |
Hb_007821_030 |
0.1156145316 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 4 |
Hb_000907_180 |
0.1161519736 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001541_050 |
0.1166204836 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 6 |
Hb_000237_200 |
0.1174234398 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 7 |
Hb_012506_060 |
0.118228149 |
- |
- |
hypothetical protein CICLE_v10029628mg [Citrus clementina] |
| 8 |
Hb_021374_030 |
0.119051064 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 9 |
Hb_065525_080 |
0.1197051548 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 10 |
Hb_000086_080 |
0.1203310682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_077549_010 |
0.1204244215 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 12 |
Hb_155215_020 |
0.1208214244 |
- |
- |
PREDICTED: RNA-binding protein pno1-like [Populus euphratica] |
| 13 |
Hb_000061_180 |
0.1213961925 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 14 |
Hb_001769_170 |
0.1250477898 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 15 |
Hb_029351_010 |
0.1265858116 |
- |
- |
PREDICTED: HD domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002157_120 |
0.1268639267 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000840_200 |
0.1272305645 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase small chain, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_157023_020 |
0.1286489855 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000212_120 |
0.1294721461 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 20 |
Hb_002471_250 |
0.1296831123 |
- |
- |
Tapt1/CMV receptor, putative isoform 2 [Theobroma cacao] |