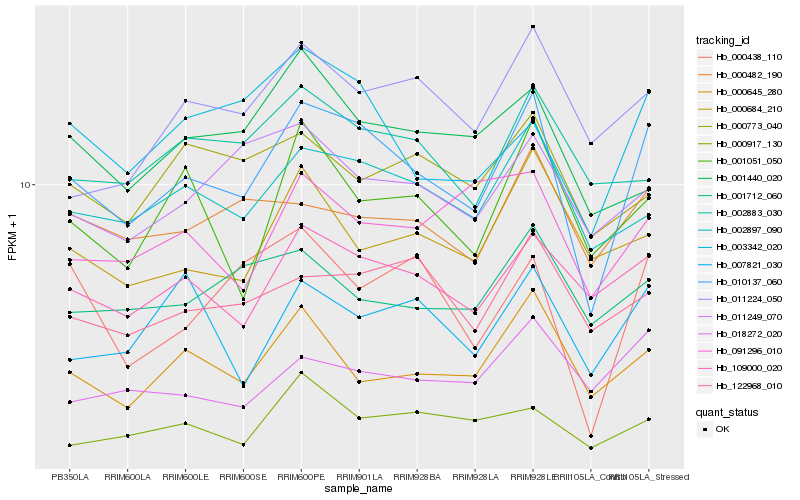
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010137_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634409 [Jatropha curcas] |
| 2 |
Hb_002897_090 |
0.1093746632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_109000_020 |
0.1136561881 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 4 |
Hb_003342_020 |
0.121625011 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 5 |
Hb_000645_280 |
0.1227903576 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 6 |
Hb_122968_010 |
0.1258240531 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 7 |
Hb_000482_190 |
0.126466163 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 8 |
Hb_001051_050 |
0.1289830323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011224_050 |
0.131257984 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Populus euphratica] |
| 10 |
Hb_011249_070 |
0.1312855559 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 11 |
Hb_007821_030 |
0.1335828114 |
- |
- |
5'-nucleotidase domain-containing [Gossypium arboreum] |
| 12 |
Hb_000438_110 |
0.1338483666 |
- |
- |
PREDICTED: adenine/guanine permease AZG1 [Jatropha curcas] |
| 13 |
Hb_000684_210 |
0.1342850526 |
- |
- |
PREDICTED: uncharacterized protein LOC105642177 [Jatropha curcas] |
| 14 |
Hb_001712_060 |
0.1345179359 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 15 |
Hb_002883_030 |
0.1349112088 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 16 |
Hb_018272_020 |
0.1353060382 |
- |
- |
PREDICTED: small glutamine-rich tetratricopeptide repeat-containing protein alpha [Jatropha curcas] |
| 17 |
Hb_091296_010 |
0.1361186902 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 18 |
Hb_000917_130 |
0.1368707521 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 19 |
Hb_000773_040 |
0.138135216 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001440_020 |
0.1383617161 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |