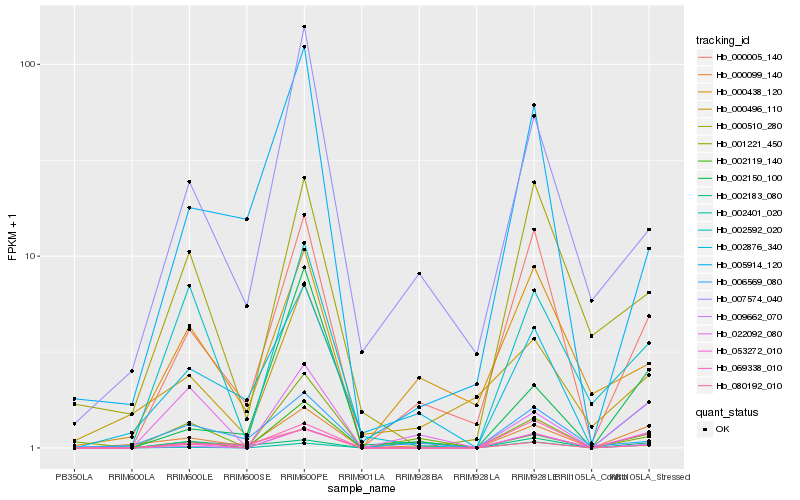
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_140 |
0.0 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 2 |
Hb_053272_010 |
0.2012109788 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 3 |
Hb_000005_140 |
0.2071140581 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa] |
| 4 |
Hb_069338_010 |
0.2180214993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002119_140 |
0.2268680625 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 6 |
Hb_002876_340 |
0.2422582356 |
- |
- |
hypothetical protein POPTR_0013s11770g [Populus trichocarpa] |
| 7 |
Hb_022092_080 |
0.253729609 |
- |
- |
- |
| 8 |
Hb_000496_110 |
0.254488419 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 9 |
Hb_009662_070 |
0.2646738814 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 10 |
Hb_007574_040 |
0.2685703698 |
- |
- |
PREDICTED: uncharacterized protein LOC105638144 [Jatropha curcas] |
| 11 |
Hb_006569_080 |
0.2734006519 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein VITISV_024559 [Vitis vinifera] |
| 12 |
Hb_002592_020 |
0.2763732052 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002150_100 |
0.2764436112 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Jatropha curcas] |
| 14 |
Hb_005914_120 |
0.2804270408 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 15 |
Hb_000438_120 |
0.2806011314 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 16 |
Hb_002183_080 |
0.2839897587 |
- |
- |
catalytic, putative [Ricinus communis] |
| 17 |
Hb_002401_020 |
0.2851795412 |
- |
- |
PREDICTED: protein argonaute 16 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000510_280 |
0.2876771115 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like [Jatropha curcas] |
| 19 |
Hb_080192_010 |
0.291680189 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_001221_450 |
0.2929649209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |